tibble(Value = 0:15,
pmf=c(0,0,1,2,3,4,5,6,5,4,3,2,1,0,0,0)/36) |>
ggplot(aes(Value,pmf)) + geom_line() + geom_point() +
theme_minimal(base_size = 20)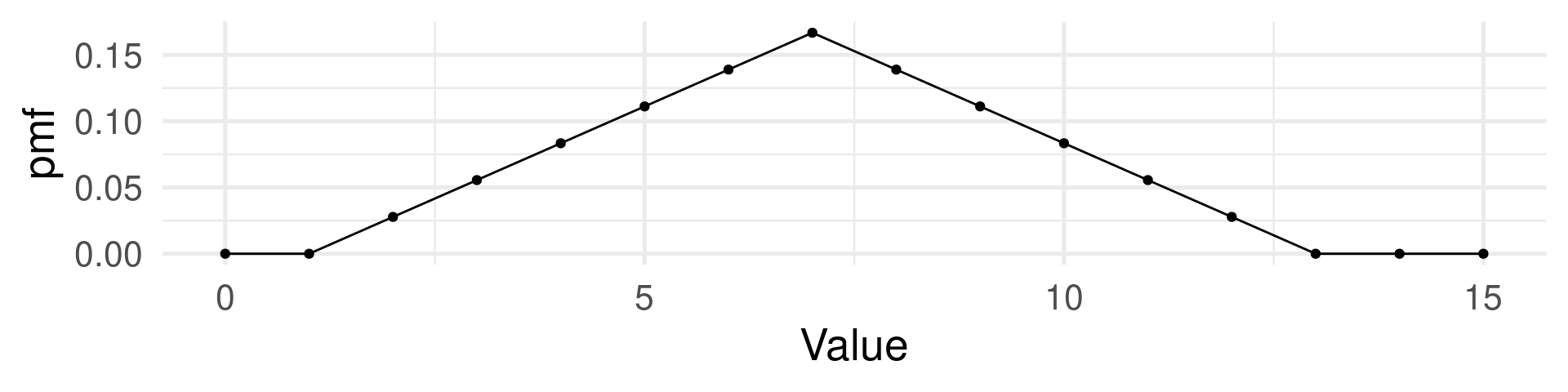
In statistics (that means, in data):
A theoretical assumption about a numerical variable in a data frame.
In probability (that means, in theory):
A mapping of events (subsets of a sample space of atomic events) to numerical values.
Why?
For random variables we can:
A random variable is
Together with a probability function \(\text{Pr}: \mathcal{F}(S)\to [0,1]\) probabilities can be assigned to values of the random variable (see the probability mass function in explained later).
3 random processes spaces and an example of a random variable \(X\):
Two coin tosses:
Sample space: \(\{HH, HT, TH, TT\}\)
We define \(X\) as the number of HEADS.
Values of \(X\): \(HH \to 2\), \(HT \to 1\), \(TH \to 1\), and \(TT \to 0\).
62 randomly selected organ donations:
Sample space: All possibilities to select 62 organ donations
We define \(X\) to be the number of complications (compare Hypothesis Testing material).
Values of \(X\): 0, 1, 2,…, 61, 62
Select a random palmer penguin:
We define \(X\) as its flipper length.
Sample space: All penguins in dataset
Values of \(X\): flipper_length_mm
Summary: A random variable is a way to look at a numerical aspect of a sample space. It often simplfies because more atomic events may be mapped to the same number.
A random variable \(X\) can be either
Discrete: \(X\) can take only a finite number of possible numeric values. Or:
Continuous: \(X\) can take values from an infinite set of real numbers (for example an interval or all positive real numbers)
Are these random variables discrete or continuous?
For working with data: Every data frame is finite, so every random variable built on data variable dataset is technically discrete. However, it can make sense to assume it as continuous because of its continuous nature. (In variables of continuous nature, many or all values are unique.)
For
the probability mass function \(f_X: \mathbb{R} \to [0,1]\) is defined as
\[f_X(x) = \text{Pr}(X=x),\]
where \(\text{Pr}(X=x)\) is an abbreviation for \(\text{Pr}(\{a\in S\text{ for which } X(a) = x\})\).
Two coin tosses \(S = \{HH, HT, TH, TT\}\)
Random variable: The sum of both dice.
Events: All 36 combinations of rolls 1+1, 1+2, 1+3, 1+4, 1+5, 1+6, 2+1, 2+2, 2+3, 2+4, 2+5, 2+6, 3+1, 3+2, 3+3, 3+4, 3+5, 3+6, 4+1, 4+2, 4+3, 4+4, 4+5, 4+6, 5+1, 5+2, 5+3, 5+4, 5+5, 5+6, 6+1, 6+2, 6+3, 6+4, 6+5, 6+6
Possible values of the random variable: 2, 3, 4, 5, 6, 7, 8, 9, 10, 11, 12 (These are numbers.)
Probability mass function: (Assuming each number has probability of \(\frac{1}{6}\) for each die.)
\(\text{Pr}(2) = \text{Pr}(12) = \frac{1}{36}\)
\(\text{Pr}(3) = \text{Pr}(11) = \frac{2}{36}\)
\(\text{Pr}(4) = \text{Pr}(10) = \frac{3}{36}\)
\(\text{Pr}(5) = \text{Pr}(9) = \frac{4}{36}\)
\(\text{Pr}(6) = \text{Pr}(8) = \frac{5}{36}\)
\(\text{Pr}(7) = \frac{6}{36}\)
The expected value of \(X\) is defined as \(E(X) = \sum_{i=1}^k p_i x_i = p_1x_1 + \dots + p_kx_k.\)
Think of \(E(X)\) as the probability weighted arithmetic mean of all possible values of \(X\).
Examples: \(X\) one roll of a die 🎲.
\(E(X) = 1\cdot\frac{1}{6} + 2\cdot\frac{1}{6} + 3\cdot\frac{1}{6} + 4\cdot\frac{1}{6} + 5\cdot\frac{1}{6} + 6\cdot\frac{1}{6} = \frac{21}{6} = 3.5\)
\(X\) sum of two die rolls 🎲🎲.
\(E(X) = 2\cdot\frac{1}{36} + 3\cdot\frac{2}{36} + 4\cdot\frac{3}{36} + 5\cdot\frac{4}{36} + \dots + 10\cdot\frac{3}{36} + 11\cdot\frac{2}{36} + 12\cdot\frac{1}{36} = 7\)
\(X\) flipper length in penguins from Palmer penguins: \(E(X) = 200.9152047\)
mean(penguins$flipper_length_mm, na.rm = TRUE)
| Type of Random Variable | Table | Graph | Formula |
|---|---|---|---|
| Discrete | ✓ | ✓ | ✓ |
| Continuous | ✗ | ✓ | ✓ |
The probability mass function \(f_X\) defines the distribution of a random variable \(X\) that is discrete.
The probability density function \(f_X\) defines the distribution of a random variable \(X\) that is continuous. (Details later)
For a column in a data frame the distribution is typically visualized by
Discrete
Technically island is cannot be interpreted as a random variable because its values are not numbers, but we also speak about the distribution of the variables.
With automatic bandwidth (bw)
A first example for a theoretical random variable.
The number of HEADS in several coin tosses and the number of complications in randomly selected organ donations are examples of random variable which have a binomial distribution.
Definition:
The binomial distribution with parameters \(n\) and \(p\) is
the number of successes in a sequence of \(n\) independent Bernoulli trials
which each delivers a success with probability \(p\) and a failure with probability \((1-p)\).
\[f(k,n,p) = \Pr(k;n,p) = \Pr(X = k) = \binom{n}{k}p^k(1-p)^{n-k}\]
where \(k\) is the number of successes, \(n\) is the number of Bernoulli trials, and \(p\) the success probability.
Probability to have exactly 3 complications in 62 randomly selected organ donations with complication probability \(p=0.1\) is
The probability to have 3 complications or less can be computed as
This was the p-value we computed with simulation for the hypothesis testing example.
For \(X \sim \text{Binom}(n,p)\) (read “\(X\) has a binomial distribution with samplesize \(n\) and success probability \(p\)”)
The expected value of \(X\) is by definition
\[E(X) = \underbrace{\sum_{k = 0}^n k}_{\text{sum over successes}} \cdot \underbrace{\binom{n}{k}p^k(1-p)^{n-k}}_{\text{probability of successes}}\]
Computation shows that \(E(X) = p\cdot n\).
Example: For \(n = 62\) organ donations with complication probability \(p=0.1\), the expected number of complications is \(E(X) = 6.2\).
Compute the p-value:
[1] 0.001455578 0.010027317 0.033981465 0.075514366[1] 0.1209787Plotting the probability mass function
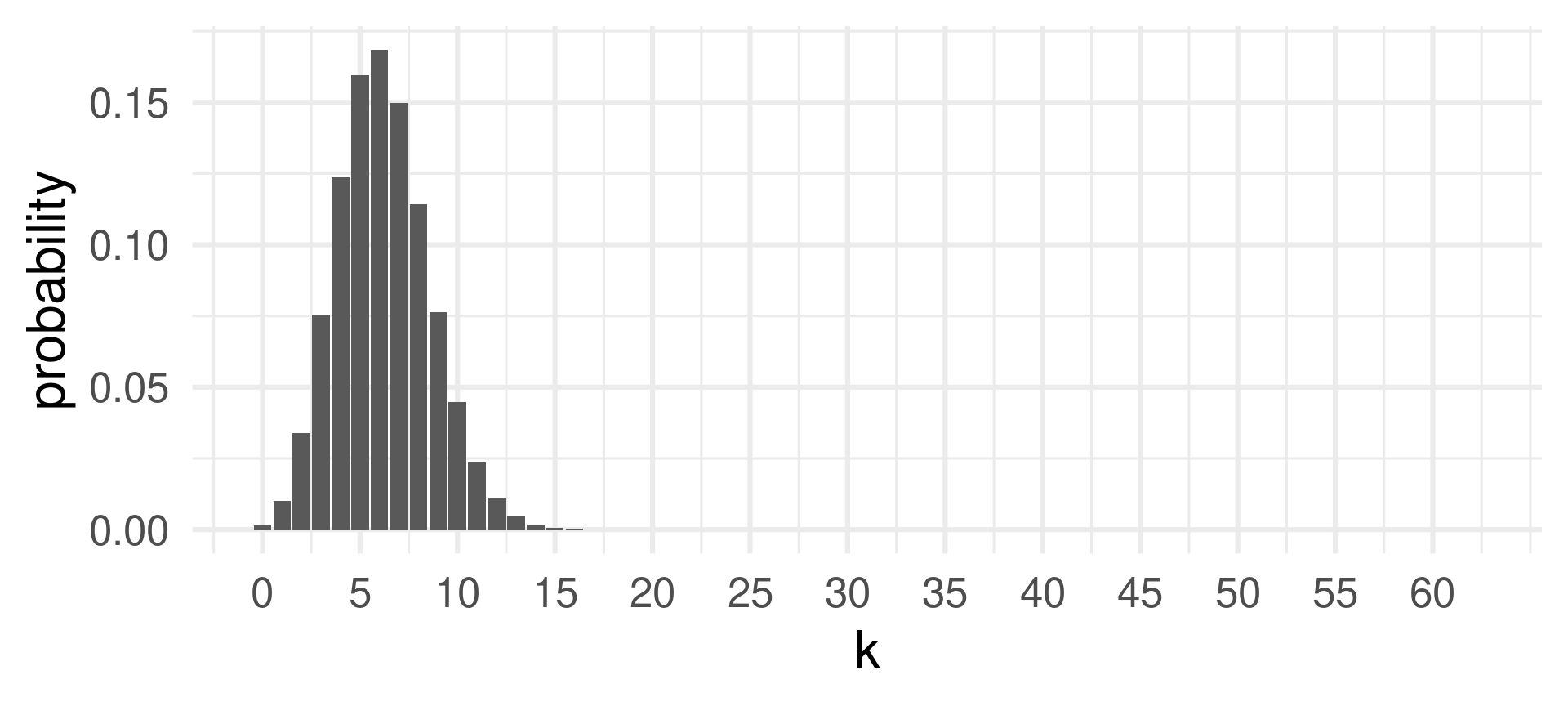
See that the highest probability is achieved for \(k=6\) which is close to the expected value of successes \(E(X) = 6.2\) for \(X \sim \text{Binom}(62,0.1)\).
Changing the sample size \(n\) when the success probability \(p = 0.1\) and the number of successes \(k=3\) is fixed:
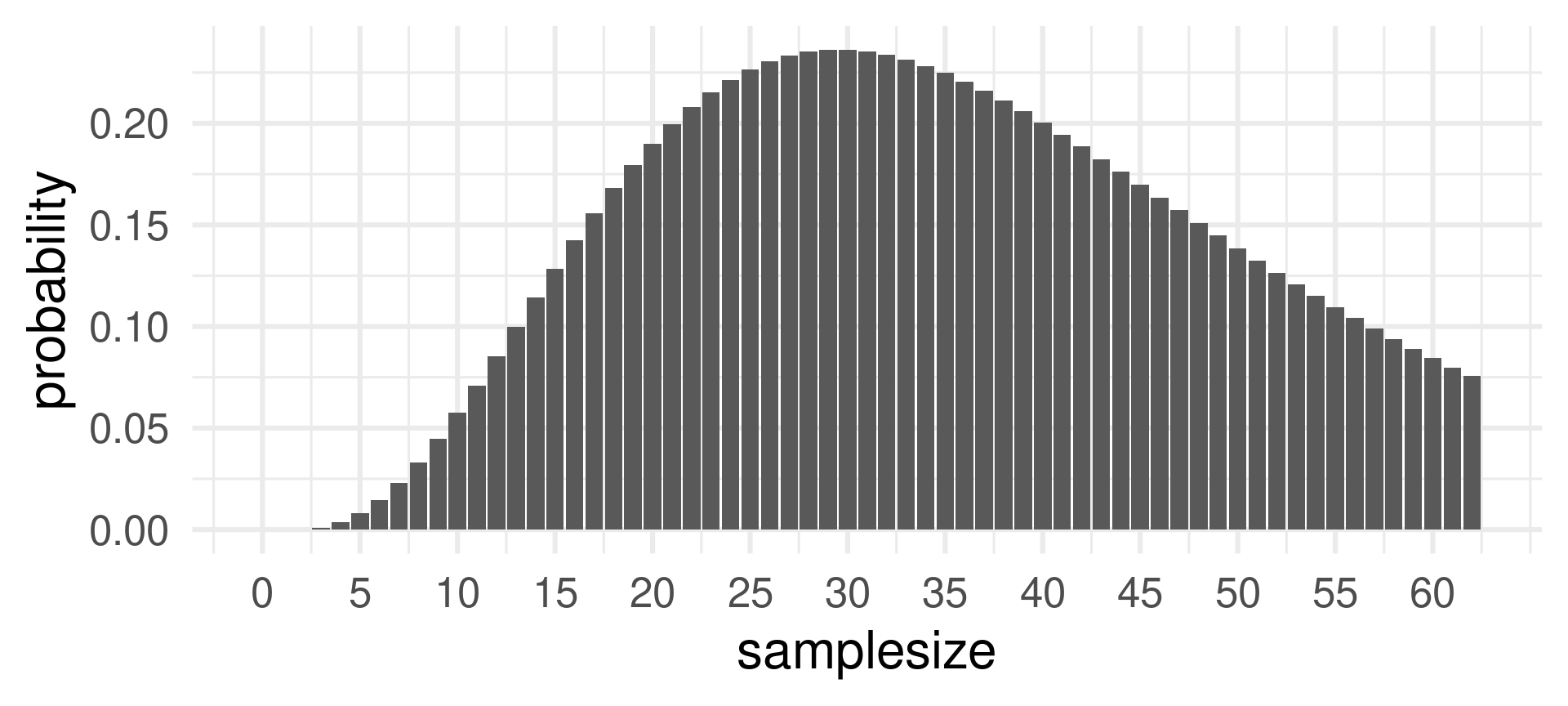
The probability of 3 successes is most likely for sample sizes around 30. Does is make sense?
Yes, because for \(n=30\) the expected value for probability \(p=0.1\) is \(3 = pn = 0.1\cdot 30\).
Changing the sample size \(p\) when the sample size \(n = 62\) and the number of successes \(k=3\) is fixed:
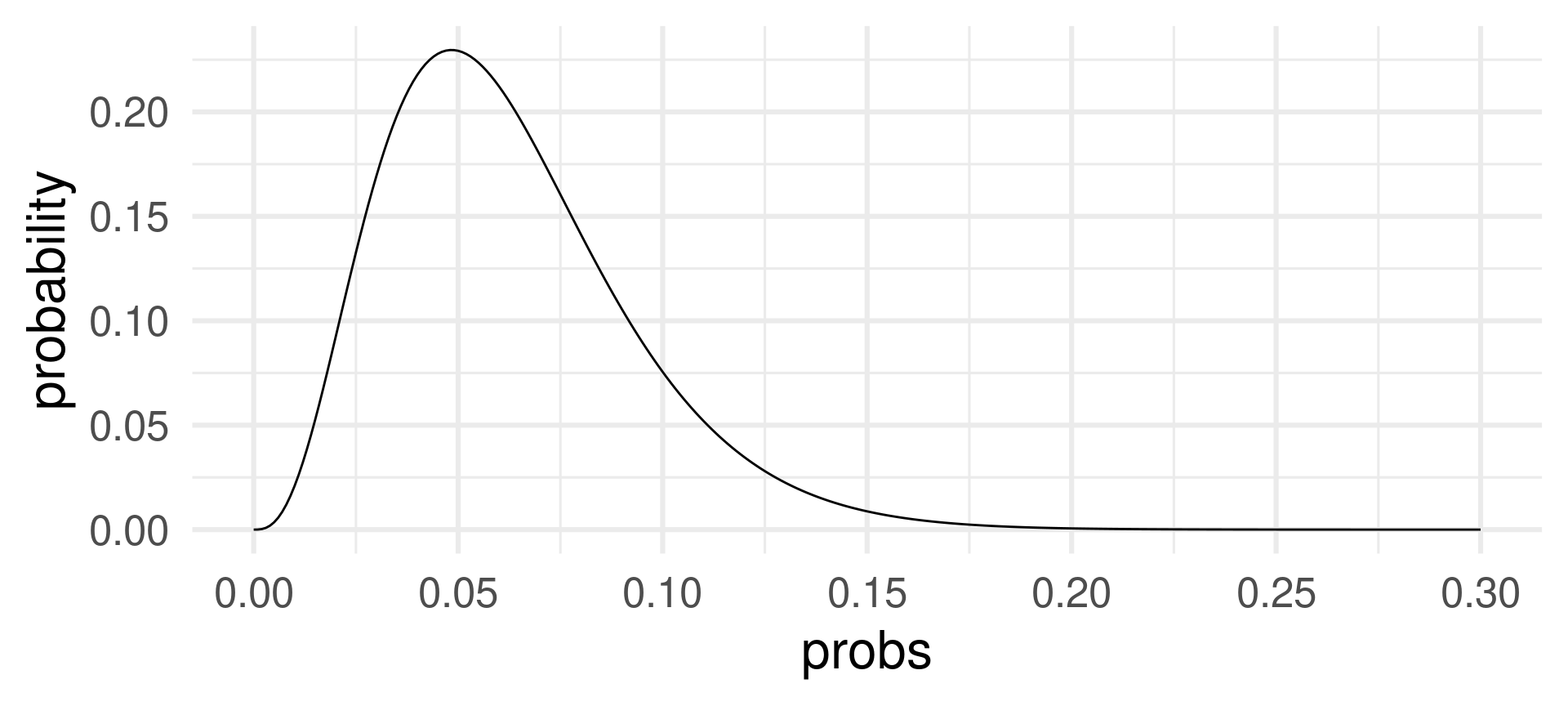
The probability of 3 successes in 62 draws is most likely for success probabilities around 0.05.
For \(p=0.05\) the expected value for \(n=62\) is \(pn = 0.05\cdot 62 = 3.1\).
In R we usually have 4 function for each distribution: The d, p, q, and r version. For the binomial distribution:
dbinom the density function (more on the name later)
pbinom distribution function
qbinom the quantile function, and
rbinom random number generator.
d…dbinom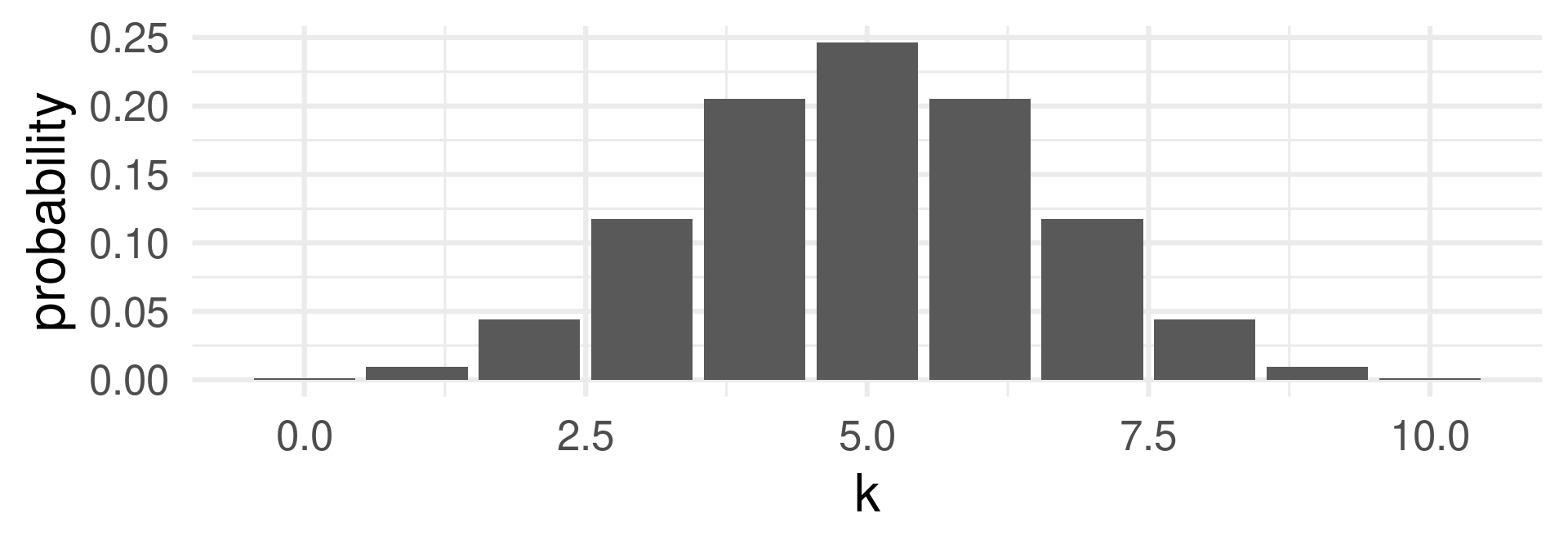
Gives the probability for the number \(x\): \(\text{Pr}(X = x)\) or \(f_X(x)\).
p…pbinom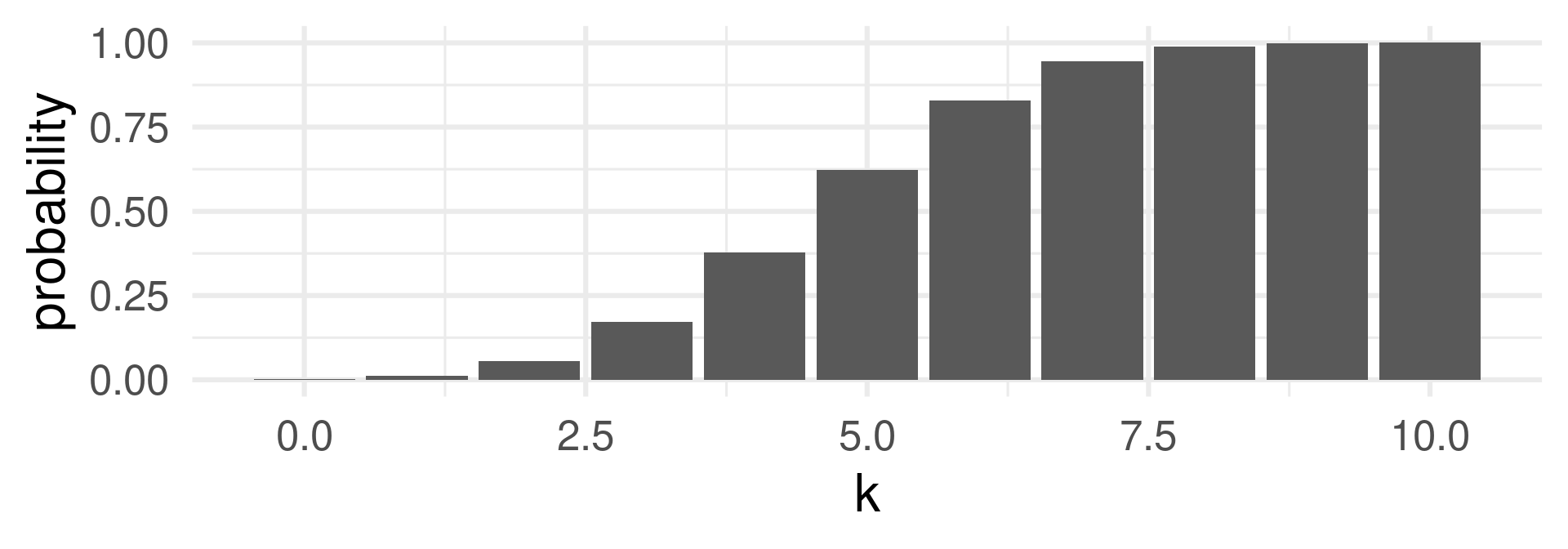
Gives the probability that the random variable is less or equal to \(x\):
\(\text{Pr}(X \leq x)\).
q…qbinom with argument \(p\) representing the fraction of lowest values of \(X\) among all values for which we want the \(k\) value for.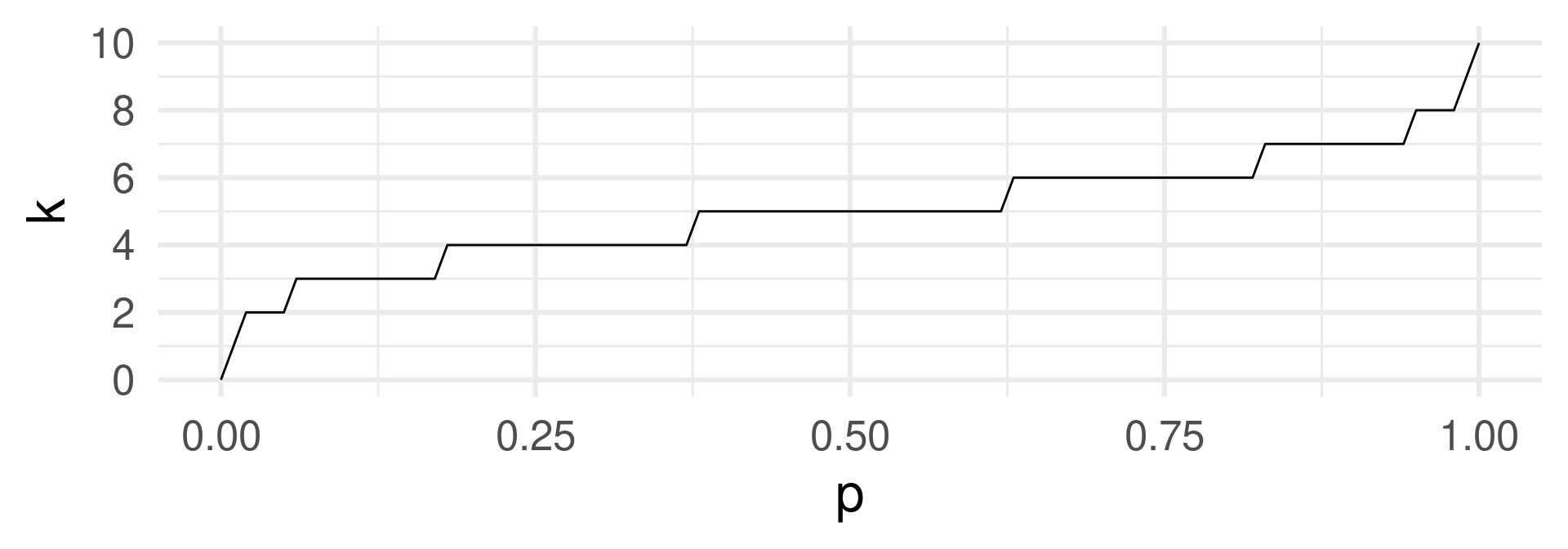
A point \((p,k)\) means: When we want a \(p\)-fraction of the probability mass, we need all events with values lower or equal to \(k\).
Quantile, distribution and mass function all carry the full information about the distribution of a random variable \(X\).
The mass function is the derivative of the distribution function.
(The distribution function is the anti-derivative of the mass function.)
[1] 0.03125 0.18750 0.50000 0.81250 0.96875 1.00000# Next comes its derivative (have to append a 0 before first)
pbinom(0:5, size = 5, prob = 0.5) |> append(0, after = 0) |> diff()[1] 0.03125 0.15625 0.31250 0.31250 0.15625 0.03125[1] 0.03125 0.15625 0.31250 0.31250 0.15625 0.03125[1] 0.03125 0.18750 0.50000 0.81250 0.96875 1.00000x and y aesthetic.probs <- seq(0, 1, by = 0.01)
k <- 0:10
q <- tibble(p = probs) |> mutate(k = qbinom(p, size = 10, prob = 0.5))
p <- tibble(k) |> mutate(p = pbinom(k, size = 10, prob = 0.5))
q_plot <- q |> ggplot(aes(p, k)) + geom_line() + theme_minimal(base_size = 24) + scale_y_continuous(breaks = seq(0, 10, 2)) + ggtitle("qbinom")
qinv_plot <- q |> ggplot(aes(k, p)) + geom_line() + theme_minimal(base_size = 24) + scale_x_continuous(breaks = seq(0, 10, 2)) + ggtitle("qbinom inverse")
p_plot <- p |> ggplot(aes(k, p)) + geom_col() + theme_minimal(base_size = 24) + scale_x_continuous(breaks = seq(0, 10, 2)) + ggtitle("pbinom")
pinv_plot <- p |> ggplot(aes(p, k)) + geom_col(orientation = "y") + theme_minimal(base_size = 24) + scale_y_continuous(breaks = seq(0, 10, 2)) + ggtitle("pbinom inverse")
library(patchwork)
(q_plot | p_plot) / (pinv_plot | qinv_plot)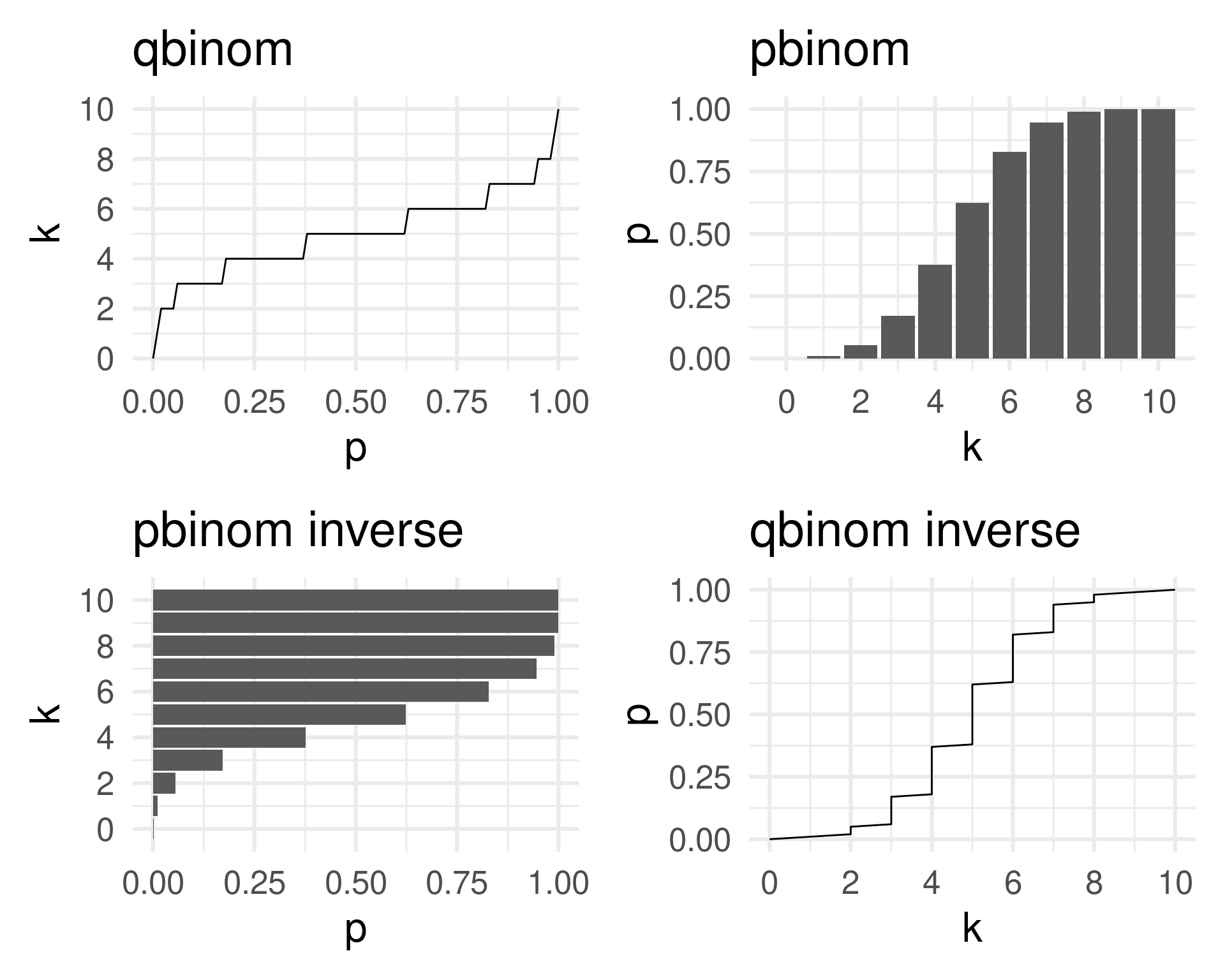
r…rbinom# 10 random binomial numbers for 62 trials with success probability 0.1
rbinom(10, size = 62, prob = 0.1) [1] 8 7 6 7 6 9 7 4 5 4# A tibble: 11 × 3
euftf n prob
<dbl> <int> <dbl>
1 0 3361 0.0736
2 1 1787 0.0391
3 2 2830 0.0620
4 3 3586 0.0786
5 4 3739 0.0819
6 5 10286 0.225
7 6 4589 0.101
8 7 5165 0.113
9 8 4692 0.103
10 9 1786 0.0391
11 10 3826 0.0838Mass function and distribution function
eu_mass <- eu |> ggplot(aes(euftf, prob)) + geom_col() + theme_minimal(base_size = 24) + scale_x_continuous(breaks = seq(0, 10, 2))
eu_distr <- eu |> mutate(cumprob = cumsum(prob)) |>
ggplot(aes(euftf, cumprob)) + geom_col() + theme_minimal(base_size = 24) + scale_x_continuous(breaks = seq(0, 10, 2))
eu_mass | eu_distr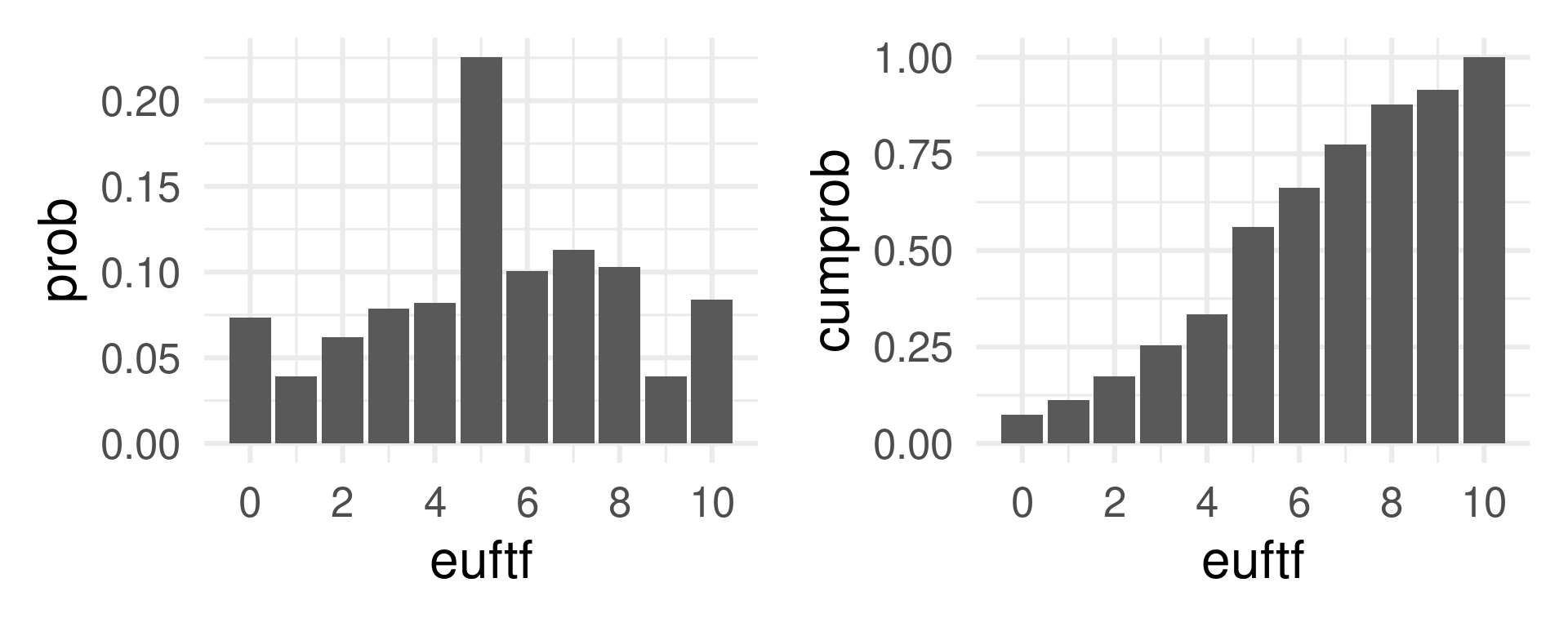
Discrete random variable
Atomic event: 20 (unfair) coin flips with HEADS probability 40%.
Random Variable: Number of HEADS.
Binomial distribution function
Continuous random variable
Atomic event: Point on a ruler of 1 meter length. Each point is equally likely.
Random Variable: The marking on the ruler in meters (number from 0 to 1).
Uniform distribution function
Interpret a point of these graphs: \(y\)-value is the probability of the event \(X \leq x\).
Corresponding mass functions:
Discrete random variable
Atomic event: 20 (unfair) coin flips with HEADS probability 40%.
Random Variable: Number of HEADS.
Binomial density function
Note: A more formal treatment of density functions on later slides!
Discrete random variable
Atomic event: Ask a European about attitude towards the EU.
Random Variable: The answer on the scale 0 to 10.
Continuous random variable
Atomic event: A visitors estimates the weight of the meat of an ox.
Random Variable: The estimated value converted to pounds.
Empirical data is always finite, so why bother with theoretical continuous distributions?
For discrete random variables the probability mass function gives us the probabilities for each number. Mathematically it is
\(f_X(x) = \text{Pr}(X = x)\) while \(F_X(x) = \text{Pr}(X \leq x)\)
Assume the discrete values with positive probability are \(x_1 < x_2 < \dots < x_n\).
Then it is easy to see the probability mass function is the diff-function of the distribution function.
\(f_X(x_i) = F_X(x_i) - F_X(x_{i-1})\)
The distribution function of a sample of 50 random numbers from a uniform distribution.
Empirical and theoretical distribution function
::::
The distribution function of a sample of 50 random numbers from a normal distribution.
Empirical and theoretical distribution function
normal <- rnorm(50)
normal_cdf <- tibble(x = normal) %>%
arrange(x) %>% # We sort the data by size
mutate(cdf = (1:length(normal))/length(normal)) # cumulative probabilities
normal_cdf |> ggplot(aes(x, y = cdf)) + geom_step() +
geom_function(fun = pnorm, color = "red") + xlim(c(-3,3)) + theme_minimal(base_size = 24)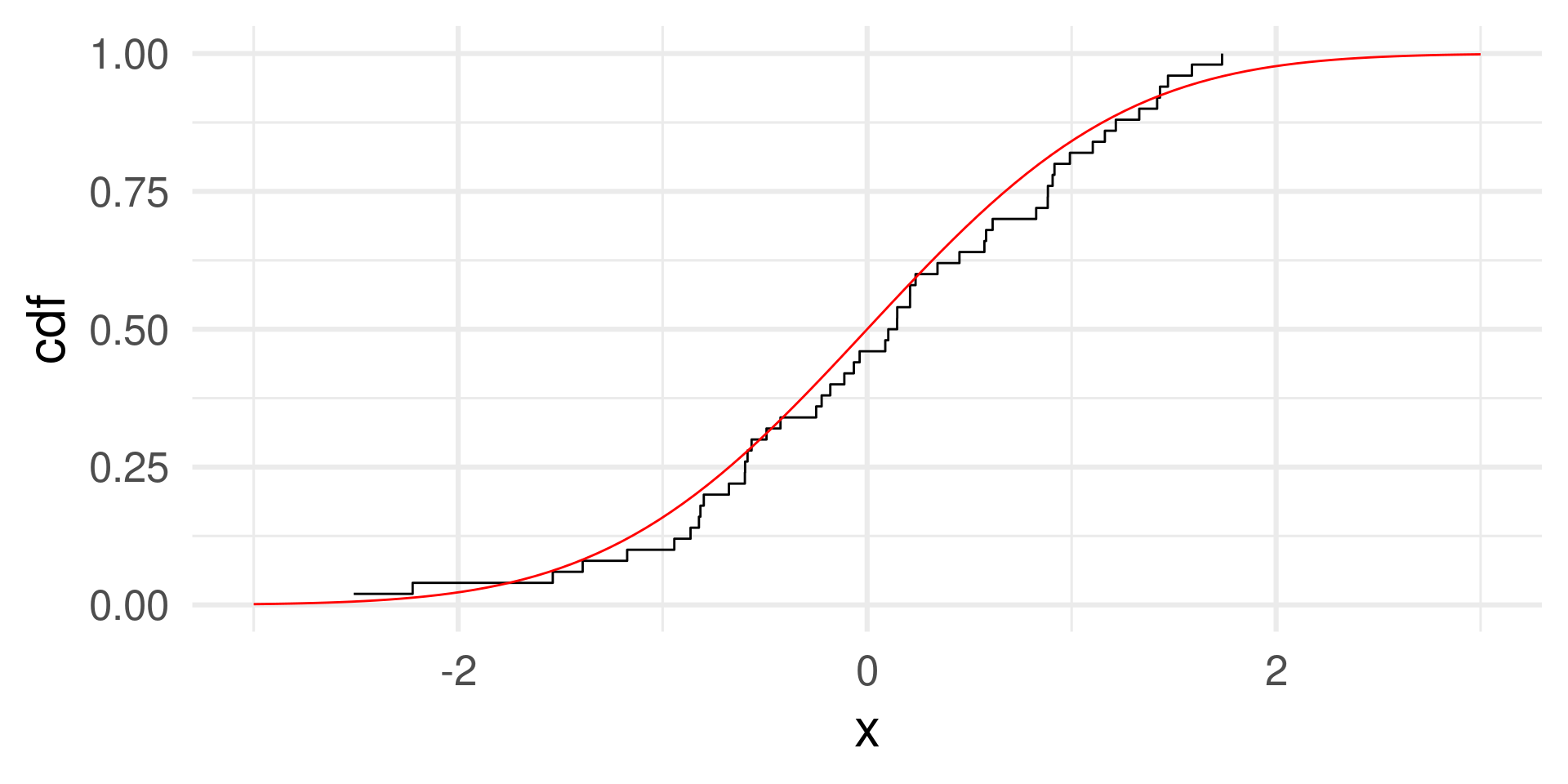
::::
The theoretical distribution is approached better with
normal <- rnorm(5000)
normal_cdf <- tibble(x = normal) %>%
arrange(x) %>% # We sort the data by size
mutate(cdf = (1:length(normal))/length(normal)) # cumulative probabilities
normal_cdf |> ggplot(aes(x, y = cdf)) + geom_step() +
geom_function(fun = pnorm, color = "red") + xlim(c(-4,4)) + theme_minimal(base_size = 24)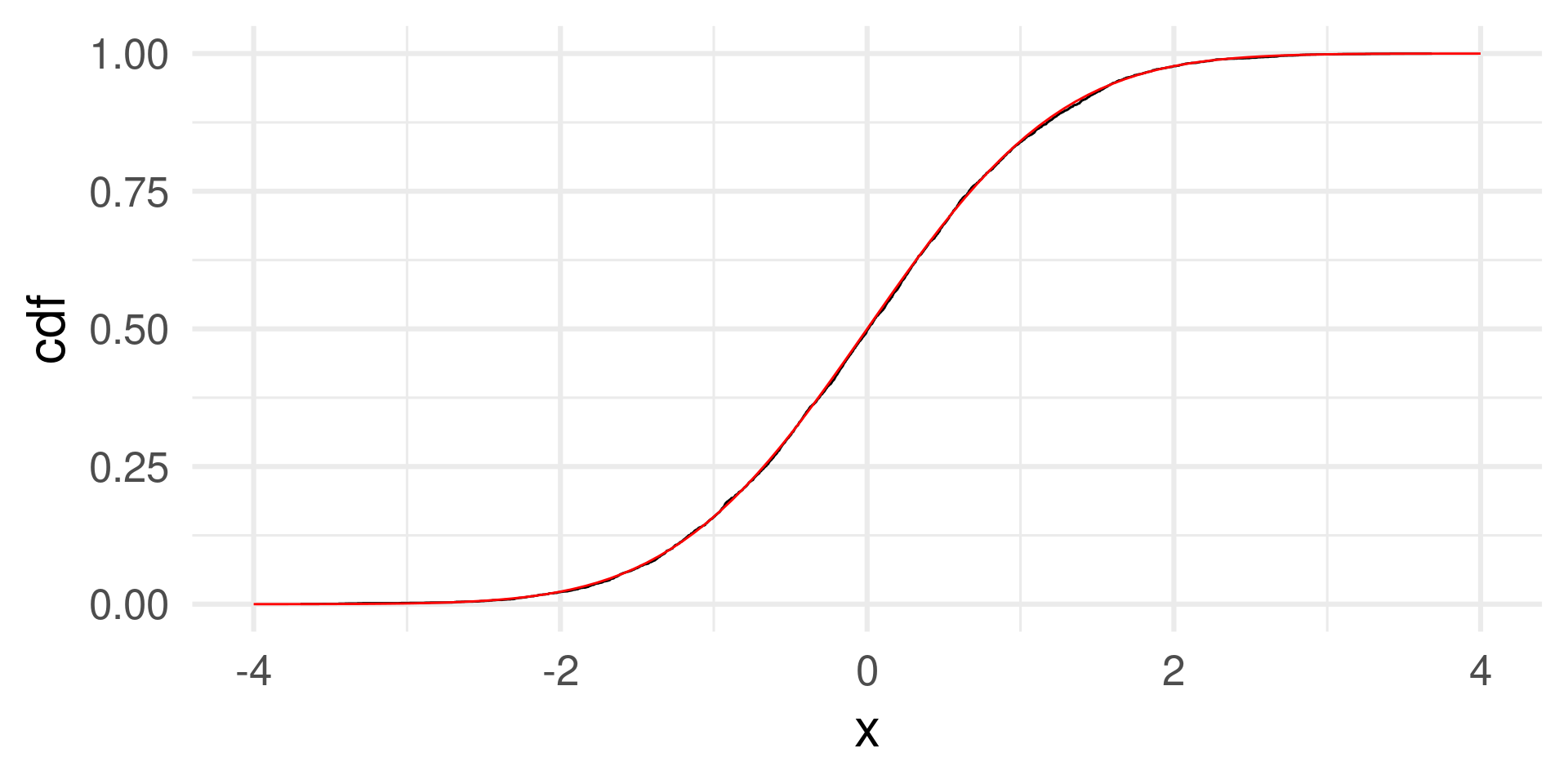
::::
The pdf is the analog of the pmf for continuous random variables.
Instead of the probability that \(X\) takes a specific value \(a\), we are interested in the probability that \(X\) takes a value in an interval \([a,b]\).
Identifiers for distributions:
unif uniform distribution
norm normal distribution
lnorm lognormal distribution
binom binomial distribution
As empirical samples of numbers also theoretical distributions have an expected value or mean and a variance (and a standard deviation). In theoretical distributions they often become (related to) parameters of the distribution.
The normal distribution has the parameters mean and sd
ggplot() +
geom_function(fun = function(x) dnorm(x, mean = 2, sd = 1)) +
geom_function(fun = function(x) dnorm(x, mean = -3, sd = 3), color = "red") +
geom_function(fun = function(x) dnorm(x, mean = 7, sd = 0.5), color = "blue") +
geom_function(fun = function(x) dnorm(x, mean = -1, sd = 6), color = "green") +
xlim(-15,15) + theme_minimal(base_size = 24)Here are some examples of mean and standard deviation:
A normal distribution fits OK.
There are many probability distributions (implemented in R or not):
https://en.wikipedia.org/wiki/List_of_probability_distributions
With interesting relations: (

Source: https://en.wikipedia.org/wiki/Relationships_among_probability_distributions)
What does the underlying model of a normal distribution mean?
Central Limit Theorem (colloquial version) The sum of many independent random variables (which can have various distributions) approaches the normal distribution (for ever larger sums and proper normalization).
\(Y = X_1 + X_2 + \dots + X_n \to \text{Normal distribution}\) for large \(n\)
n <- 10000
tibble(X1 = runif(n),X2 = runif(n),X3 = runif(n),X4 = runif(n),X5 = runif(n),
X6 = runif(n),X7 = runif(n),X8 = runif(n),X9 = runif(n)) %>%
mutate(S2 = X1 + X2,
S5 = X1 + X2 + X3 + X4 + X5,
S9 = X1 + X2 + X3 + X4 + X5 + X6 + X7 + X8 + X9 ) %>%
ggplot() +
geom_histogram(aes(x = X1, y =..density..), binwidth = 0.1, alpha = 0.5) +
geom_histogram(aes(x = S2, y =..density..), binwidth = 0.1, fill = "green", alpha = 0.5) +
geom_histogram(aes(x = S5, y =..density..), binwidth = 0.1, fill = "red", alpha = 0.5) +
geom_histogram(aes(x = S9, y =..density..), binwidth = 0.1, fill = "blue", alpha = 0.5) + xlim(c(-0.5,9)) Why are sums of random variables important?
What when \(Y\) is the product of many positive-valued random variable?
\(Y = X_1 \cdot X_2 \cdot \dots \cdot X_n\)
Then
\(\log(Y) = \log(X_1) + \log(X_2) + \dots + \log(X_n) \to \text{Normal distribution}\) for large \(n\) (central limit theorem)
So, \(\log(Y)\) tends to become normally distributed.
\(Y\) is called to have a lognormal distribution.
n <- 10000
g <- tibble(X1 = 2*runif(n),X2 = 2*runif(n),X3 = 2*runif(n),X4 = 2*runif(n),X5 = 2*runif(n),
X6 = 2*runif(n),X7 = 2*runif(n),X8 = 2*runif(n),X9 = 2*runif(n)) %>%
mutate(S2 = X1 * X2,
S5 = X1 * X2 * X3 * X4 * X5,
S9 = X1 * X2 * X3 * X4 * X5 * X6 * X7 * X8 * X9 ) %>%
ggplot() +
geom_histogram(aes(x = X1, y =..density..), binwidth = 0.1, alpha = 0.5) +
geom_histogram(aes(x = S2, y =..density..), binwidth = 0.1, fill = "green", alpha = 0.5) +
geom_histogram(aes(x = S5, y =..density..), binwidth = 0.1, fill = "red", alpha = 0.5) +
geom_histogram(aes(x = S9, y =..density..), binwidth = 0.1, fill = "blue", alpha = 0.5) + xlim(c(-0.5,9))
gDistributions “live” on different domains. This determines which values for random numbers are theoretically possible.
Take away:
For many variables there are better or worse candidates for a theoretical distribution to assume.
