ggplot() +
geom_function(fun = function(x) log(x/(1-x)), xlim=c(0.001,0.999), n = 500) +
geom_function(fun = function(x) 1/(1 + exp(-x)), color = "red") +
scale_x_continuous(breaks = seq(-5,5,1), limits = c(-5,5)) +
scale_y_continuous(breaks = seq(-5,5,1), limits = c(-5,5)) +
coord_fixed() + theme_minimal(base_size = 24) + labs(x = "x")W#08: Logistic Regression, Classification Problems, Prediction
Logistic regression
Logistic regression
- Logistic regression is a GLM used to model a binary categorical outcome using numerical and categorical predictors.
- The distribution is the Bernoulli distribution with parameter \(p\) (success probability).
- As link function connecting \(\eta_i\) to \(p_i\) we use the logit function.
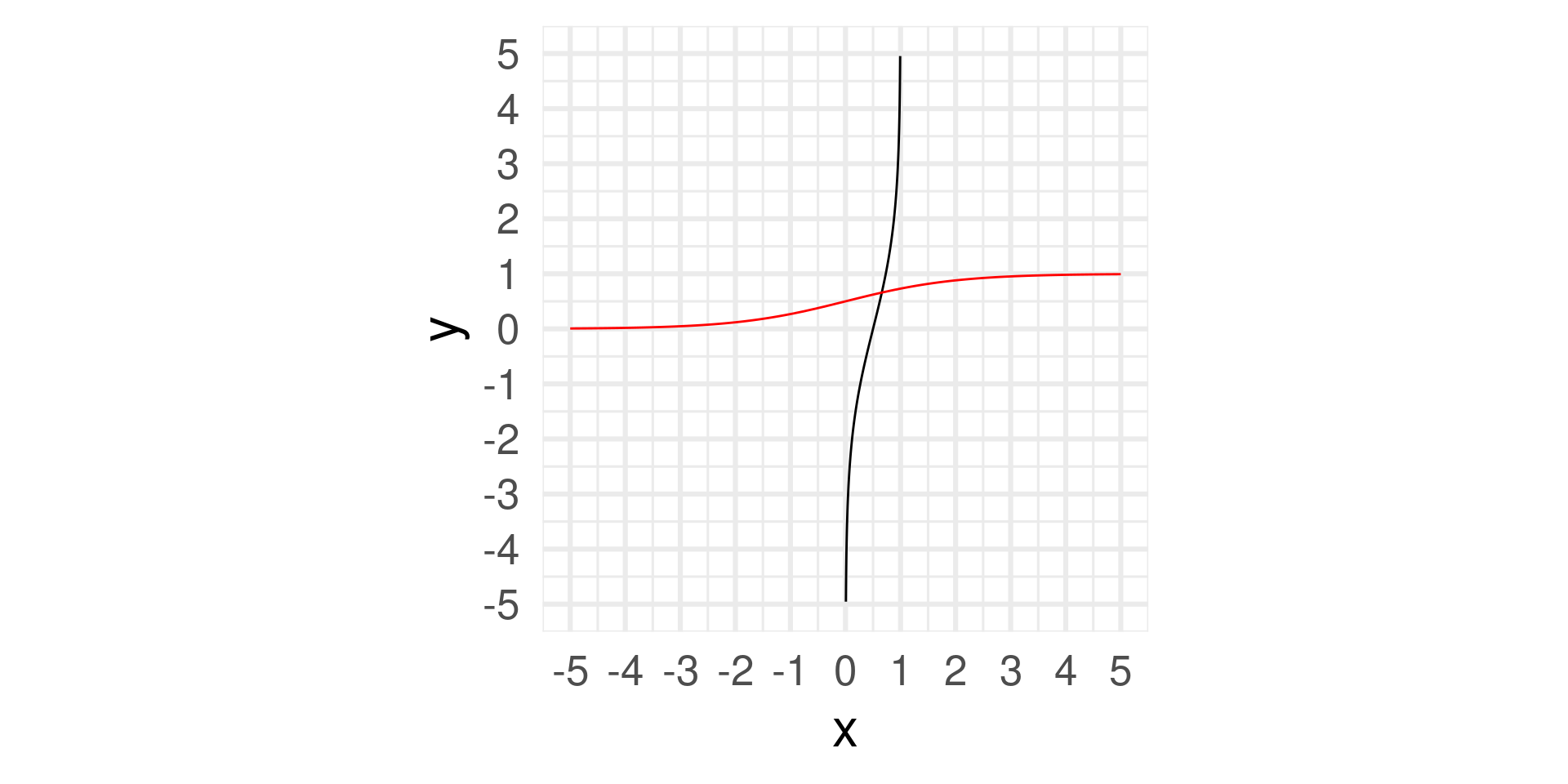
- Logit function: \(\text{logit}: [0,1] \to \mathbb{R}\)
\[\text{logit}(p) = \log\left(\frac{p}{1-p}\right)\]
- \(\frac{p}{1-p}\) is called the odds of a success which happens with probability \(p\).
Example: Roll a six with a die has \(p=1/6\). Thus, the odds are \(\frac{1/6}{5/6} = 1/5\). Sometimes written as 1:5. “The odds of success are one to five.”
Properties of the logit
- Logit takes values between 0 and 1 and returns values between \(-\infty\) and \(\infty\)
- The inverse of the logit function if the logistic function (mapping values from \(-\infty\) and \(\infty\) to values between 0 and 1): \[\text{logit}^{-1}(x) = \text{logistic}(x) = \frac{e^x}{1+e^x} = \frac{1}{1+e^{-x}}\]
- Logit can be interpreted as the log odds of a success – more on this later.
Logit and logistic function
The logistic regression model
- Based on the three GLM criteria we have
- \(y_i \sim \text{Bernoulli}(p_i)\)
- \(\eta_i = \beta_0+\beta_1 x_{1,i} + \cdots + \beta_n x_{n,i}\)
- \(\text{logit}(p_i) = \eta_i\)
- From which we get
\[p_i = \frac{e^{\beta_0+\beta_1 x_{1,i} + \cdots + \beta_k x_{k,i}}}{1 + e^{\beta_0+\beta_1 x_{1,i} + \cdots + \beta_k x_{k,i}}}\]
Modeling spam
With tidymodels we fit a GLM in the same way as a linear model except we
- specify the model with
logistic_reg() - use
"glm"instead of"lm"as the engine - define
family = "binomial"for the link function to be used in the model
Spam model
# A tibble: 2 × 5
term estimate std.error statistic p.value
<chr> <dbl> <dbl> <dbl> <dbl>
1 (Intercept) -1.80 0.0716 -25.1 2.04e-139
2 num_char -0.0621 0.00801 -7.75 9.50e- 15Model: \[\log\left(\frac{p}{1-p}\right) = -1.80-0.0621\cdot \text{num_char}\]
Predicted probability: Examples
We can compute the predicted probability that an email with 2000 character is spam as follows:
\[\log\left(\frac{p}{1-p}\right) = -1.80-0.0621\cdot 2 = -1.9242\]
(Note: num_char is in thousands.)
\[\frac{p}{1-p} = e^{-1.9242} = 0.15 \Rightarrow p = 0.15 \cdot (1 - p)\]
\[p = 0.15 - 0.15\cdot p \Rightarrow 1.15\cdot p = 0.15\]
\[p = 0.15 / 1.15 = 0.13\]
Predicted probability
logistic <- function(t) 1/(1+exp(-t))
preds <- tibble(x=c(2,15,40), y = logistic(-1.80-0.0621*x))
email |> ggplot(aes(x = num_char, y = as.numeric(spam)-1)) +
geom_point(alpha = 0.2) +
geom_function(fun = function(x) logistic(-1.80-0.0621*x),color="red") +
geom_point(data = preds, mapping = aes(x,y), color = "blue", size = 3)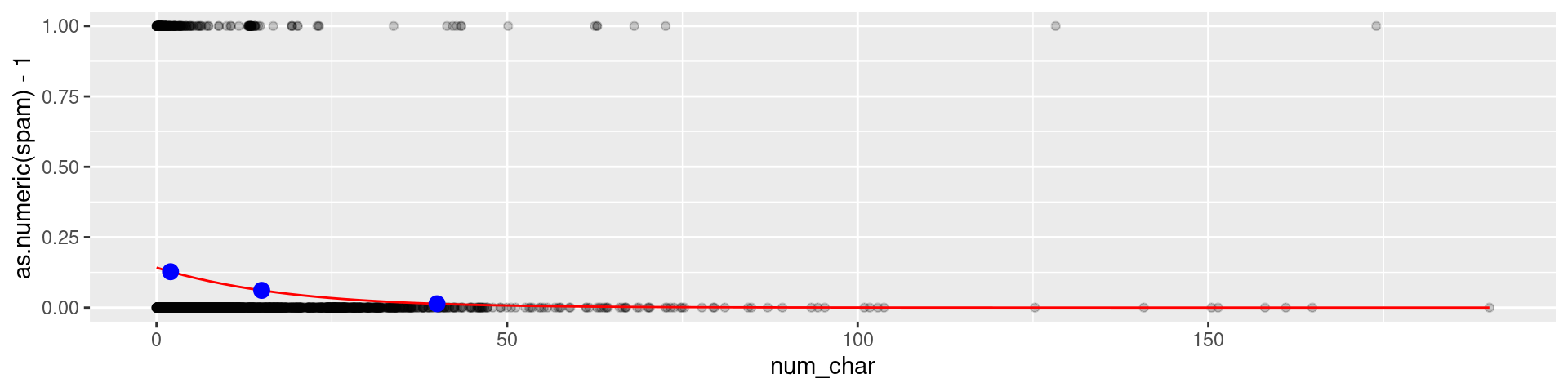
Spam probability 2,000 characters: 0.1273939
Spam probability 15,000 characters: 0.06114
Spam probability 40,000 characters: 0.0135999
Interpretation of coefficients
\[\log\left(\frac{p}{1-p}\right) = -1.80-0.0621\cdot \text{num_char}\]
What does an increase by thousand characters (num_char + 1) imply?
Let us assume the predicted probability of an email is \(p_0\). Then an increase of num_char by one implied that the log-odds become
\[\log\left(\frac{p_0}{1-p_0}\right) - 0.0621 = \log\left(\frac{p_0}{1-p_0}\right) - \log(e^{0.0621})\]
\[ = \log\left(\frac{p_0}{1-p_0}\right) + \log(\frac{1}{e^{0.0621}}) = \log\left(\frac{p_0}{1-p_0} \frac{1}{e^{0.0621}}\right) = \log\left(\frac{p_0}{1-p_0} 0.94\right)\]
That means the odds of being spam decrease by 6%.
Penguins example
library(palmerpenguins)
sex_fit <- logistic_reg() |>
set_engine("glm") |>
fit(sex ~ body_mass_g, data = na.omit(penguins), family = "binomial")
tidy(sex_fit)# A tibble: 2 × 5
term estimate std.error statistic p.value
<chr> <dbl> <dbl> <dbl> <dbl>
1 (Intercept) -5.16 0.724 -7.13 1.03e-12
2 body_mass_g 0.00124 0.000173 7.18 7.10e-13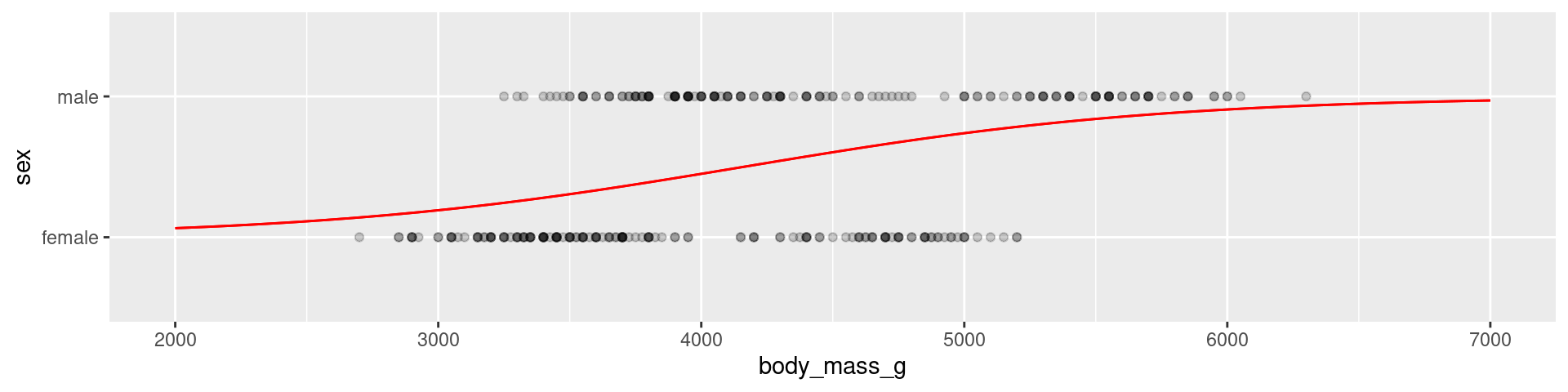
Summarizing the Idea of Logistic Regression
- We want to explain/predict a binary response variable.
- We want to use the concept of linear models.
- To that end we first interprete each outcome as the outcome of a probabilisitic event happening with probability \(p_i\). That way we transform the problem from modeling a binary outcome to a numerical outcome.
- However, probabilities are bounded between 0 and 1.
- We use the logit function to map probabilities to the real line. That means we predict the log-odds of the probability.
- We model the log-odds with a linear model.
Sensitivity and specificity
False positive and negative
| Email labelled spam | Email labelled not spam | |
|---|---|---|
| Email is spam | True positive | False negative (Type 2 error) |
| Email is not spam | False positive (Type 1 error) | True negative |
Confusion matrix
More general: Confusion matrix of statistical classification:
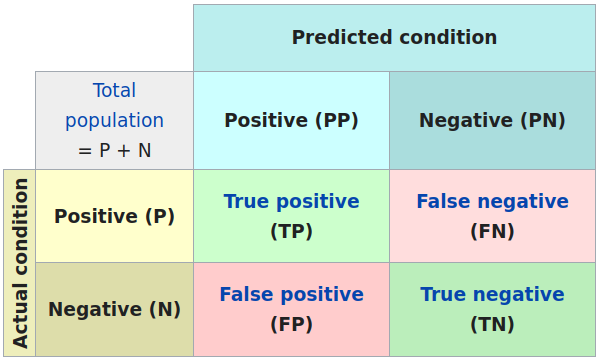
Sensitivity and specificity

Sensitivity is the true positive rate: TP / (TP + FN)
Specificity is the true negative rate: TN / (TN + FP)
For spam a positive case is an email labelled as spam:
Sensitivity: Fraction of emails labelled as spam among all emails which are spam.
Low sensitivity \(\to\) More false negatives \(\to\) More spam in you inbox!
Specificity: Fraction of emails labelled as not spam among all emails which are not spam.
Low specificity \(\to\) More false positives \(\to\) More relevant emails in spam folder!
If you were designing a spam filter, would you want sensitivity and specificity to be high or low? What are the trade-offs associated with each decision?
COVID-19 tests
A COVID-19 test is called positive when it indicates the presence of the virus.
What is the sensitivity of a test?
Probability the test is positive when the tested person has COVID-19. (The true positive rate.)
What is the specificity of a test?
Probability the test is negative when the tested person does not have COVID-19. (The true negative rate.)
Often the sensitivity is around 90% and the specificity is around 99%. What does that mean?
- When you do not have COVID the test is more likely to be correct, than when you do have COVID. However, specificity and sensitivity do not tell something about the probability to have COVID when the test is positive, or to not have COVID when the test is negative.
Another view
Prediction
Prediction vs. Explanation
Explanation and Prediction are different Modelling Mindsets1.
- With an Explanatory Mindset we want to understand the relationship between the predictors and the outcome.
- We interpret coefficients
- We ask what variance can be explained by a linear model or a PCA
- With a Predictive Mindset we want to predict the outcome for new observations.
- We use the model to predict the outcome for new observations
- We ask how well we can predict the outcome for new observations
- We may ignore why the model works
Important: The mindsets are very different but they may use the same models!
Prediction Example: Building a spam filter
- Data: Set of emails and we know
- if each email is spam or not
- several other features
- Use logistic regression to predict the probability that an incoming email is spam
- Use model selection to pick the model with the best predictive performance
- Building a model to predict the probability that an email is spam is only half of the battle! We also need a decision rule about which emails get flagged as spam (e.g. what probability should we use as out cutoff?)
- A simple approach: choose a single threshold probability and any email that exceeds that probability is flagged as spam
Emails: Use all predictors
logistic_reg() |>
set_engine("glm") |>
fit(spam ~ ., data = email, family = "binomial") |>
tidy() |> print(n = 22)Warning: glm.fit: fitted probabilities numerically 0 or 1 occurred# A tibble: 22 × 5
term estimate std.error statistic p.value
<chr> <dbl> <dbl> <dbl> <dbl>
1 (Intercept) -9.09e+1 9.80e+3 -0.00928 9.93e- 1
2 to_multiple1 -2.68e+0 3.27e-1 -8.21 2.25e-16
3 from1 -2.19e+1 9.80e+3 -0.00224 9.98e- 1
4 cc 1.88e-2 2.20e-2 0.855 3.93e- 1
5 sent_email1 -2.07e+1 3.87e+2 -0.0536 9.57e- 1
6 time 8.48e-8 2.85e-8 2.98 2.92e- 3
7 image -1.78e+0 5.95e-1 -3.00 2.73e- 3
8 attach 7.35e-1 1.44e-1 5.09 3.61e- 7
9 dollar -6.85e-2 2.64e-2 -2.59 9.64e- 3
10 winneryes 2.07e+0 3.65e-1 5.67 1.41e- 8
11 inherit 3.15e-1 1.56e-1 2.02 4.32e- 2
12 viagra 2.84e+0 2.22e+3 0.00128 9.99e- 1
13 password -8.54e-1 2.97e-1 -2.88 4.03e- 3
14 num_char 5.06e-2 2.38e-2 2.13 3.35e- 2
15 line_breaks -5.49e-3 1.35e-3 -4.06 4.91e- 5
16 format1 -6.14e-1 1.49e-1 -4.14 3.53e- 5
17 re_subj1 -1.64e+0 3.86e-1 -4.25 2.16e- 5
18 exclaim_subj 1.42e-1 2.43e-1 0.585 5.58e- 1
19 urgent_subj1 3.88e+0 1.32e+0 2.95 3.18e- 3
20 exclaim_mess 1.08e-2 1.81e-3 5.98 2.23e- 9
21 numbersmall -1.19e+0 1.54e-1 -7.74 9.62e-15
22 numberbig -2.95e-1 2.20e-1 -1.34 1.79e- 1The prediction task
- The mechanics of prediction is easy:
- Plug in values of predictors to the model equation
- Calculate the predicted value of the response variable, \(\hat{y}\)
- Getting it right is harder
- There is no guarantee the model estimates you have are correct
- Or that your model will perform as well with new data as it did with your sample data
Balance Over- and Underfitting
In a one predictor model we can show both visually:
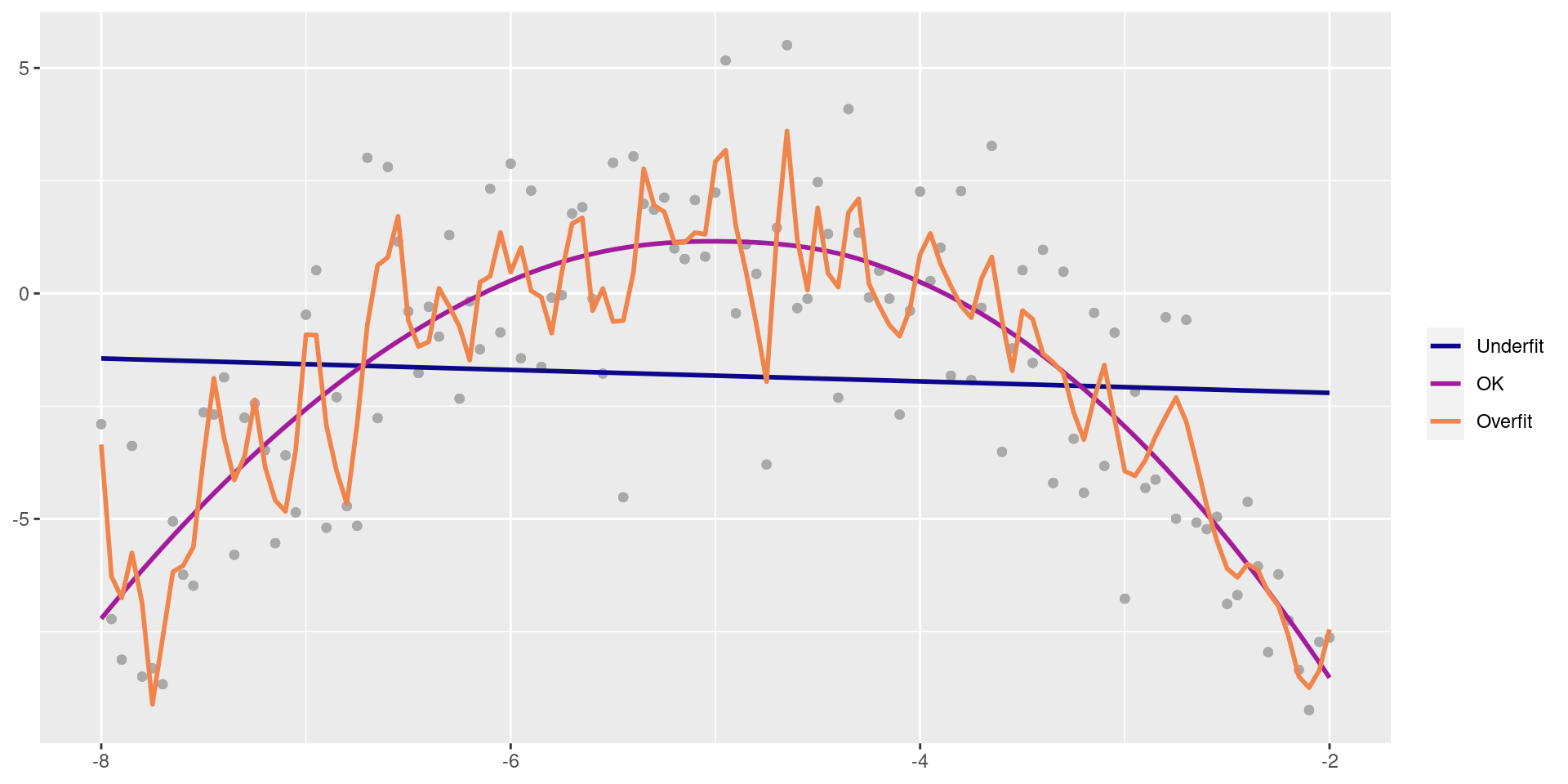
Spending our data
Problem:
- There are several steps to create a useful prediction model: parameter estimation, model selection, performance assessment, etc.
- Doing all of this on the entire data we have available can lead to overfitting.
Solution: We subsets our data for different tasks, as opposed to allocating all data to parameter estimation (as we have done so far).
Splitting Data
Splitting data
- Training set:
- Sandbox for model building
- Spend most of your time using the training set to develop the model
- Majority of the data (usually 80%)
- Testing set:
- Held in reserve to determine efficacy of one or two chosen models
- Critical to look at it once, otherwise it becomes part of the modeling process
- Remainder of the data (usually 20%)
Sidenote: Splitting data is very important for the predictive questions, because we want to know how well our model performs on new data. It is not relevant for explanatory questions where we want to interpret coefficients and infer about the underlying population.
Performing the split
# Fix random numbers by setting the seed
# Enables analysis to be reproducible when random numbers are used
set.seed(1116)
# Put 80% of the data into the training set
email_split <- initial_split(email, prop = 0.80)
# Create data frames for the two sets:
train_data <- training(email_split)
test_data <- testing(email_split)A note on the seed: The seed is an arbitrary number that is used to initialize a pseudorandom number generator. The same seed will always result in the same sequence of pseudorandom numbers. This is useful for reproducibility.
Peek at the split
Rows: 3,136
Columns: 21
$ spam <fct> 0, 1, 0, 1, 0, 0, 0, 0, 0, 0, 1, 0, 0, 0, 0, 0, 0, 0, 0, …
$ to_multiple <fct> 0, 0, 0, 0, 1, 1, 0, 0, 0, 0, 0, 0, 0, 1, 0, 0, 0, 0, 0, …
$ from <fct> 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, …
$ cc <int> 2, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 35, 0, 0, 0, 0, 0,…
$ sent_email <fct> 1, 0, 1, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 1, 0, 0, 0, 0, …
$ time <dttm> 2012-01-25 23:46:55, 2012-01-03 06:28:28, 2012-02-04 17:…
$ image <dbl> 0, 0, 0, 0, 0, 1, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, …
$ attach <dbl> 0, 0, 0, 0, 0, 1, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, …
$ dollar <dbl> 10, 0, 0, 0, 0, 0, 13, 0, 0, 0, 2, 0, 0, 0, 14, 0, 0, 0, …
$ winner <fct> no, no, no, no, no, no, no, yes, no, no, no, no, no, no, …
$ inherit <dbl> 0, 0, 0, 0, 0, 1, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, …
$ viagra <dbl> 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, …
$ password <dbl> 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 3, 0, 0, …
$ num_char <dbl> 23.308, 1.162, 4.732, 42.238, 1.228, 25.599, 16.764, 10.7…
$ line_breaks <int> 477, 2, 127, 712, 30, 674, 367, 226, 98, 671, 46, 192, 67…
$ format <fct> 1, 0, 1, 1, 0, 1, 1, 1, 1, 1, 0, 1, 0, 0, 1, 1, 1, 1, 1, …
$ re_subj <fct> 1, 0, 1, 0, 0, 0, 0, 0, 0, 0, 0, 0, 1, 0, 1, 0, 0, 0, 1, …
$ exclaim_subj <dbl> 0, 0, 0, 0, 0, 0, 1, 0, 0, 0, 1, 0, 0, 0, 0, 0, 0, 0, 1, …
$ urgent_subj <fct> 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, …
$ exclaim_mess <dbl> 12, 0, 2, 2, 2, 31, 2, 0, 0, 1, 0, 1, 2, 0, 2, 0, 11, 1, …
$ number <fct> small, none, big, big, small, small, small, small, small,…Rows: 785
Columns: 21
$ spam <fct> 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, …
$ to_multiple <fct> 1, 0, 0, 0, 0, 0, 0, 0, 0, 0, 1, 0, 0, 1, 0, 0, 0, 0, 0, …
$ from <fct> 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, …
$ cc <int> 0, 1, 0, 1, 4, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 3, 0, …
$ sent_email <fct> 1, 1, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 1, 0, …
$ time <dttm> 2012-01-01 18:55:06, 2012-01-01 20:38:32, 2012-01-02 06:…
$ image <dbl> 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 1, 0, 0, …
$ attach <dbl> 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 1, 0, 0, 1, 0, 0, …
$ dollar <dbl> 0, 0, 5, 0, 0, 0, 0, 5, 4, 0, 0, 0, 21, 0, 0, 2, 9, 0, 0,…
$ winner <fct> no, no, no, no, no, no, no, no, no, no, no, no, no, no, n…
$ inherit <dbl> 0, 0, 0, 0, 0, 0, 0, 0, 1, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, …
$ viagra <dbl> 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, …
$ password <dbl> 0, 0, 1, 0, 0, 0, 0, 0, 0, 1, 0, 0, 0, 0, 0, 0, 0, 0, 2, …
$ num_char <dbl> 4.837, 15.075, 18.037, 45.842, 11.438, 1.482, 14.431, 0.9…
$ line_breaks <int> 193, 354, 345, 881, 125, 24, 296, 13, 192, 14, 32, 30, 55…
$ format <fct> 1, 1, 1, 1, 0, 1, 1, 0, 1, 0, 0, 0, 1, 1, 1, 1, 1, 1, 1, …
$ re_subj <fct> 0, 1, 0, 1, 1, 0, 0, 0, 0, 0, 1, 0, 0, 0, 1, 0, 0, 1, 0, …
$ exclaim_subj <dbl> 0, 0, 1, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, …
$ urgent_subj <fct> 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, …
$ exclaim_mess <dbl> 1, 10, 20, 5, 2, 0, 0, 0, 6, 0, 0, 1, 3, 0, 4, 0, 1, 0, 1…
$ number <fct> big, small, small, big, small, none, small, small, small,…Workflow of Predictive Modeling
Fit a model to the training dataset
email_fit <- logistic_reg() |>
set_engine("glm") |>
fit(spam ~ ., data = train_data, family = "binomial")Warning: glm.fit: fitted probabilities numerically 0 or 1 occurredWe get a warning and should explore the reasons for 0 or 1 probability.
Look at categorical predictors
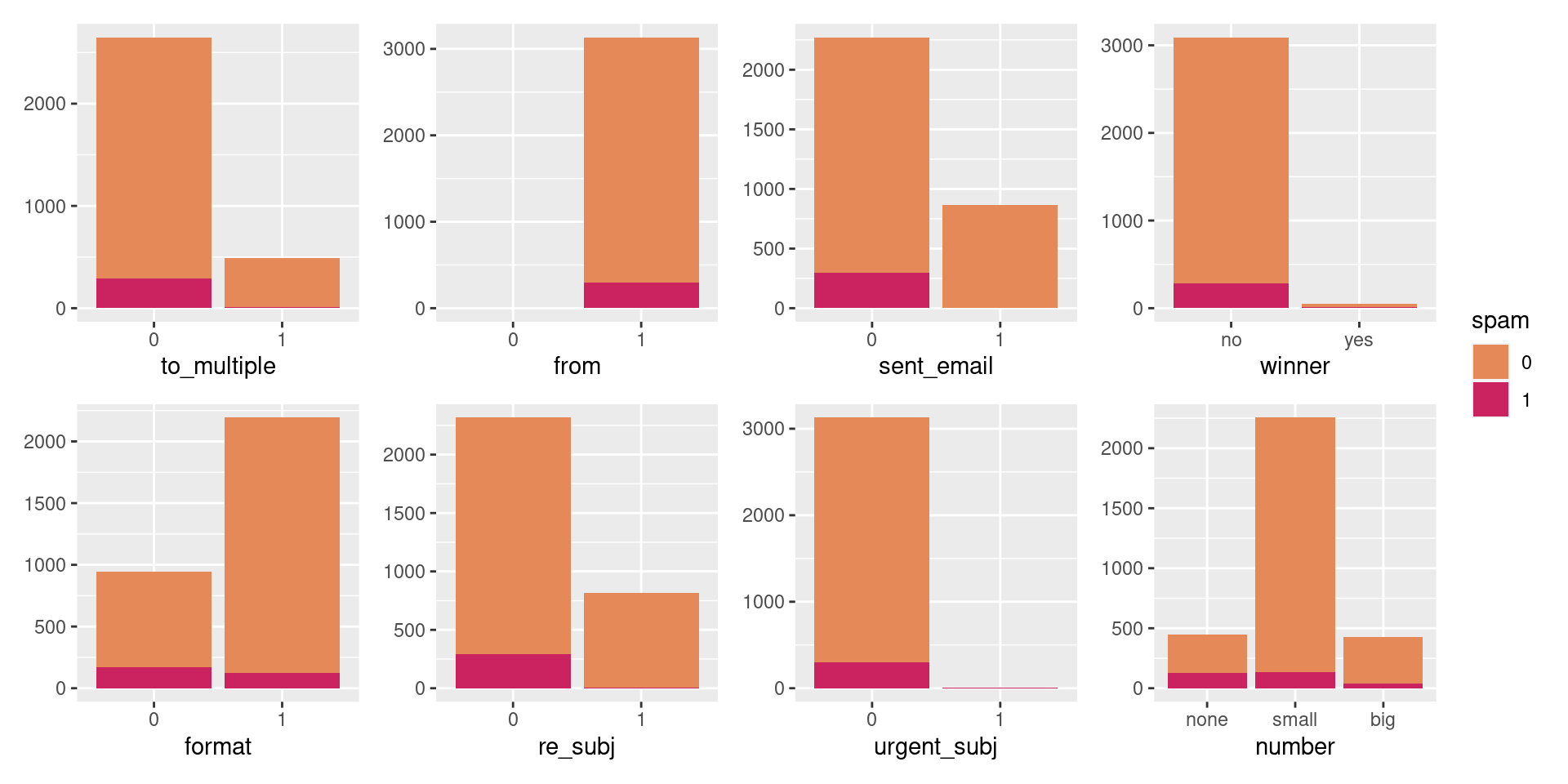
Closer look at from and sent_email.
Counting cases
from: Whether the message was listed as from anyone (this is usually set by default for regular outgoing email).
# A tibble: 3 × 3
spam from n
<fct> <fct> <int>
1 0 1 2837
2 1 0 3
3 1 1 296No non-spam mails without from.
- There is incomplete separation in the data for those variables.
- That mean we have a sure prediction probabilities (0 or 1). (That is the warning. Also, these variables have the highest coefficients.)
- This is not what we assume about reality. Maybe our sample is too small to see it.
- Therefore we exclude these variables.
Look at numerical variables
# A tibble: 22 × 4
# Groups: name [11]
name spam mean sd
<chr> <fct> <dbl> <dbl>
1 attach 0 0.124 0.775
2 attach 1 0.227 0.620
3 cc 0 0.393 2.62
4 cc 1 0.388 3.25
5 dollar 0 1.56 5.33
6 dollar 1 0.779 3.01
7 exclaim_mess 0 6.68 50.2
8 exclaim_mess 1 8.75 88.4
9 exclaim_subj 0 0.0783 0.269
10 exclaim_subj 1 0.0769 0.267
11 image 0 0.0536 0.503
12 image 1 0.00334 0.0578
13 inherit 0 0.0352 0.216
14 inherit 1 0.0702 0.554
15 line_breaks 0 247. 326.
16 line_breaks 1 108. 321.
17 num_char 0 11.4 14.9
18 num_char 1 5.63 15.7
19 password 0 0.112 0.938
20 password 1 0.0201 0.182
21 viagra 0 0 0
22 viagra 1 0.0268 0.463 viagra has no mentions in non-spam emails.
- We should exclude this variable for the same reason.
Fit a model to the training dataset
email_fit <- logistic_reg() |>
set_engine("glm") |>
fit(spam ~ . - from - sent_email - viagra, data = train_data, family = "binomial") Warning: glm.fit: fitted probabilities numerically 0 or 1 occurredparsnip model object
Call: stats::glm(formula = spam ~ . - from - sent_email - viagra, family = stats::binomial,
data = data)
Coefficients:
(Intercept) to_multiple1 cc time image
-9.867e+01 -2.505e+00 1.944e-02 7.396e-08 -2.854e+00
attach dollar winneryes inherit password
5.070e-01 -6.440e-02 2.170e+00 4.499e-01 -7.065e-01
num_char line_breaks format1 re_subj1 exclaim_subj
5.870e-02 -5.420e-03 -9.017e-01 -2.995e+00 1.002e-01
urgent_subj1 exclaim_mess numbersmall numberbig
3.572e+00 1.009e-02 -8.518e-01 -1.329e-01
Degrees of Freedom: 3135 Total (i.e. Null); 3117 Residual
Null Deviance: 1974
Residual Deviance: 1447 AIC: 1485We still get a warning, but without very high coefficients.
Predict with the testing dataset
Predicting the raw values (log-odds)
1 2 3 4 5 6
-4.942500 -6.312226 -3.938487 -6.688992 -4.399541 -1.587700 Predicting probabilities
Relate back to the model concept
email_pred <-
predict(email_fit, test_data, type = "prob") |>
select(spam_prob = .pred_1) |>
mutate(spam_logodds =
predict(email_fit, test_data, type = "raw"),
spam_odds = exp(spam_logodds)) |>
bind_cols(predict(email_fit, test_data)) |>
# Append real data
bind_cols(test_data |> select(spam))
email_pred# A tibble: 785 × 5
spam_prob spam_logodds spam_odds .pred_class spam
<dbl> <dbl> <dbl> <fct> <fct>
1 0.00709 -4.94 0.00714 0 0
2 0.00181 -6.31 0.00181 0 0
3 0.0191 -3.94 0.0195 0 0
4 0.00124 -6.69 0.00124 0 0
5 0.0121 -4.40 0.0123 0 0
6 0.170 -1.59 0.204 0 0
7 0.0410 -3.15 0.0427 0 0
8 0.139 -1.83 0.161 0 0
9 0.0617 -2.72 0.0657 0 0
10 0.0983 -2.22 0.109 0 0
# ℹ 775 more rows- The raw predictions are the log-odds.
- From which we can compute the odds.
- From which the probability is computed. Here it is done by
predict. - The
.pred_classprediction is when the probability > 0.5.- What does it mean for the odds and the log-odds?
Answers: odds > 1, log-odds > 0
Another look
# A tibble: 785 × 5
spam_prob spam_logodds spam_odds .pred_class spam
<dbl> <dbl> <dbl> <fct> <fct>
1 0.903 2.23 9.29 1 1
2 0.833 1.60 4.98 1 0
3 0.825 1.55 4.71 1 1
4 0.733 1.01 2.75 1 1
5 0.683 0.766 2.15 1 1
6 0.626 0.517 1.68 1 1
7 0.614 0.464 1.59 1 0
8 0.597 0.392 1.48 1 1
9 0.538 0.153 1.17 1 1
10 0.537 0.148 1.16 1 0
11 0.510 0.0404 1.04 1 0
12 0.491 -0.0345 0.966 0 0
13 0.490 -0.0407 0.960 0 0
14 0.489 -0.0453 0.956 0 1
15 0.483 -0.0698 0.933 0 1
16 0.473 -0.107 0.899 0 0
17 0.463 -0.150 0.861 0 0
18 0.457 -0.174 0.840 0 0
19 0.447 -0.212 0.809 0 0
20 0.447 -0.214 0.808 0 1
# ℹ 765 more rowsWe see false positives and false negatives.
Evaluate the performance
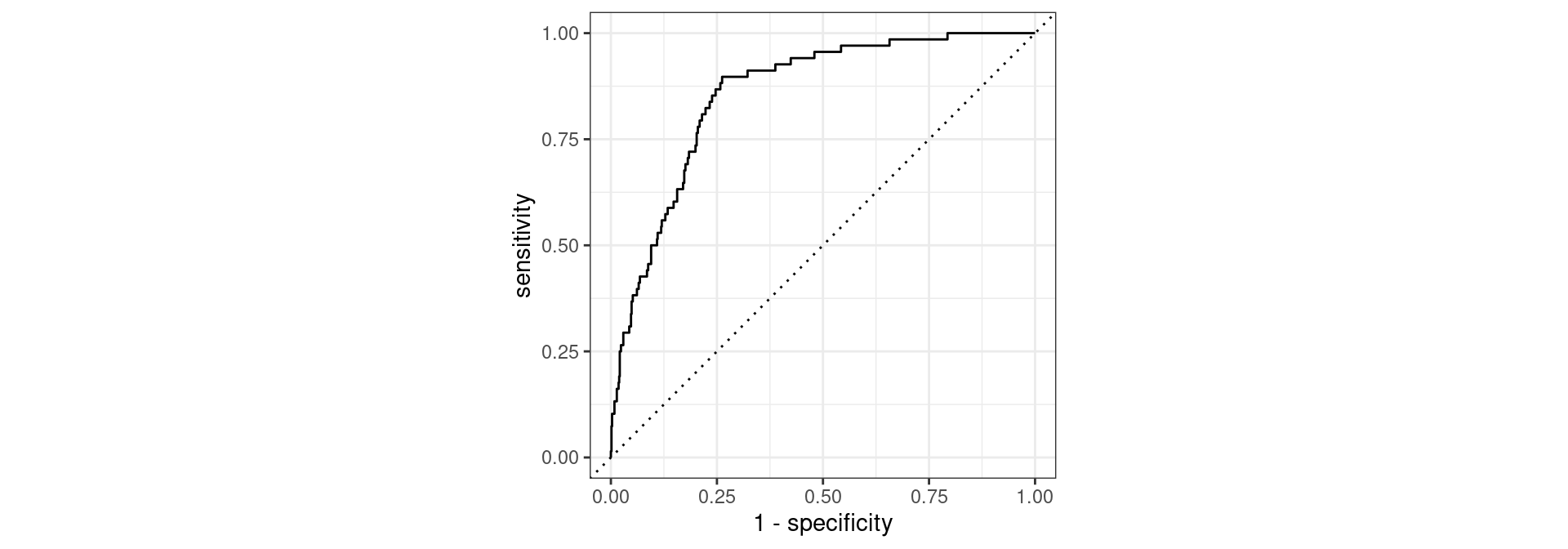
Receiver operating characteristic (ROC) curve1 which plots true positive rate (sensitivity) vs. false positive rate (1 - specificity)
Evaluate the performance
Find the area under the curve (AUC).
In calculus language:
\(\int_0^1 \text{TPR}(\text{FPR}) d\text{FPR}\)
Remember:
- TPR = True Positive Rate = sensitivity
- FPR = False Positive Rate = 1 - specificity
- specificity is the true negative rate.
Feature engineering
Feature engineering
- We prefer simple models when possible, but parsimony does not mean sacrificing accuracy (or predictive performance) in the interest of simplicity
- Variables that go into the model and how they are represented are critical to the success of the model
- Feature engineering is getting creative with our predictors in an effort to make them more useful for our model (to increase its predictive performance)
Modeling workflow, revisited
Create a recipe for feature engineering steps to be applied to the training data
- The
tidymodelsway (similar to a pipeline in python’s scikit-learn).
- The
Fit the model to the training data after these steps have been applied
Using the model estimates from the training data, predict outcomes for the test data
Evaluate the performance of the model on the test data
Recipes
Initiate a recipe
email_rec <- recipe(
spam ~ ., # formula
data = train_data # data to use for cataloguing names and types of variables
)
summary(email_rec) |> print(n = 21)# A tibble: 21 × 4
variable type role source
<chr> <list> <chr> <chr>
1 to_multiple <chr [3]> predictor original
2 from <chr [3]> predictor original
3 cc <chr [2]> predictor original
4 sent_email <chr [3]> predictor original
5 time <chr [1]> predictor original
6 image <chr [2]> predictor original
7 attach <chr [2]> predictor original
8 dollar <chr [2]> predictor original
9 winner <chr [3]> predictor original
10 inherit <chr [2]> predictor original
11 viagra <chr [2]> predictor original
12 password <chr [2]> predictor original
13 num_char <chr [2]> predictor original
14 line_breaks <chr [2]> predictor original
15 format <chr [3]> predictor original
16 re_subj <chr [3]> predictor original
17 exclaim_subj <chr [2]> predictor original
18 urgent_subj <chr [3]> predictor original
19 exclaim_mess <chr [2]> predictor original
20 number <chr [3]> predictor original
21 spam <chr [3]> outcome originalThe object email_rec only includes meta-data (columns names and types)!
Remove certain variables
Feature engineer date
- The date-time may not be such an interesting predictor.
- It could only bring in a general trend over time
- Often decomposing the date to the month or the day of the week (dow) is more interesting.
step_datecan easily extract these
Create dummy variables
- Use helper functions like
all_nominalorall_outcomesfromtidymodelsfor column selection.
Remove zero variance variables
Variables that contain only a single value.
Full recipe
email_rec <- recipe(
spam ~ ., # formula
data = train_data # data to use for cataloguing names and types of variables
) |>
step_rm(from, sent_email, viagra) |>
step_date(time, features = c("dow", "month")) |>
step_rm(time) |>
step_dummy(all_nominal(), -all_outcomes()) |>
step_zv(all_predictors())
print(email_rec)── Recipe ──────────────────────────────────────────────────────────────────────── Inputs Number of variables by roleoutcome: 1
predictor: 20── Operations • Variables removed: from, sent_email, viagra• Date features from: time• Variables removed: time• Dummy variables from: all_nominal(), -all_outcomes()• Zero variance filter on: all_predictors()The object email_rec only includes meta-data of the data frame it shall work on (a formula, columns names and types)!
Building workflows
Define model
Define workflow
Workflows bring together models and recipes so that they can be easily applied to both the training and test data.
══ Workflow ════════════════════════════════════════════════════════════════════
Preprocessor: Recipe
Model: logistic_reg()
── Preprocessor ────────────────────────────────────────────────────────────────
5 Recipe Steps
• step_rm()
• step_date()
• step_rm()
• step_dummy()
• step_zv()
── Model ───────────────────────────────────────────────────────────────────────
Logistic Regression Model Specification (classification)
Computational engine: glm Fit model to training data
# A tibble: 27 × 5
term estimate std.error statistic p.value
<chr> <dbl> <dbl> <dbl> <dbl>
1 (Intercept) -0.651 0.254 -2.57 1.03e- 2
2 cc 0.0214 0.0229 0.936 3.49e- 1
3 image -2.99 1.31 -2.28 2.28e- 2
4 attach 0.512 0.116 4.41 1.03e- 5
5 dollar -0.0651 0.0307 -2.12 3.40e- 2
6 inherit 0.440 0.205 2.15 3.14e- 2
7 password -0.723 0.302 -2.39 1.67e- 2
8 num_char 0.0585 0.0240 2.43 1.50e- 2
9 line_breaks -0.00548 0.00139 -3.94 8.24e- 5
10 exclaim_subj 0.0998 0.268 0.373 7.09e- 1
11 exclaim_mess 0.0103 0.00198 5.20 2.02e- 7
12 to_multiple_X1 -2.56 0.339 -7.56 4.11e-14
13 winner_yes 2.24 0.430 5.21 1.90e- 7
14 format_X1 -0.953 0.157 -6.06 1.38e- 9
15 re_subj_X1 -3.00 0.444 -6.76 1.39e-11
16 urgent_subj_X1 3.69 1.15 3.20 1.37e- 3
17 number_small -0.840 0.162 -5.20 1.98e- 7
18 number_big -0.0915 0.244 -0.375 7.07e- 1
19 time_dow_Mon -0.326 0.303 -1.08 2.82e- 1
20 time_dow_Tue 0.0813 0.275 0.296 7.67e- 1
21 time_dow_Wed -0.260 0.275 -0.946 3.44e- 1
22 time_dow_Thu -0.220 0.279 -0.788 4.31e- 1
23 time_dow_Fri -0.0612 0.275 -0.223 8.24e- 1
24 time_dow_Sat 0.0646 0.292 0.221 8.25e- 1
25 time_month_Feb 0.760 0.178 4.26 2.03e- 5
26 time_month_Mar 0.506 0.178 2.85 4.40e- 3
27 time_month_Apr -12.0 394. -0.0306 9.76e- 1Make predictions for test data
# A tibble: 785 × 23
.pred_0 .pred_1 spam to_multiple from cc sent_email time
<dbl> <dbl> <fct> <fct> <fct> <int> <fct> <dttm>
1 0.993 0.00653 0 1 1 0 1 2012-01-01 18:55:06
2 0.998 0.00169 0 0 1 1 1 2012-01-01 20:38:32
3 0.987 0.0127 0 0 1 0 0 2012-01-02 06:42:16
4 0.999 0.000825 0 0 1 1 0 2012-01-02 16:12:51
5 0.991 0.00876 0 0 1 4 0 2012-01-02 17:45:36
6 0.878 0.122 0 0 1 0 0 2012-01-02 22:55:03
7 0.959 0.0414 0 0 1 0 0 2012-01-03 02:07:17
8 0.852 0.148 0 0 1 0 0 2012-01-03 06:41:35
9 0.938 0.0619 0 0 1 0 0 2012-01-03 17:02:35
10 0.896 0.104 0 0 1 0 0 2012-01-03 12:14:51
# ℹ 775 more rows
# ℹ 15 more variables: image <dbl>, attach <dbl>, dollar <dbl>, winner <fct>,
# inherit <dbl>, viagra <dbl>, password <dbl>, num_char <dbl>,
# line_breaks <int>, format <fct>, re_subj <fct>, exclaim_subj <dbl>,
# urgent_subj <fct>, exclaim_mess <dbl>, number <fct>Evaluate the performance
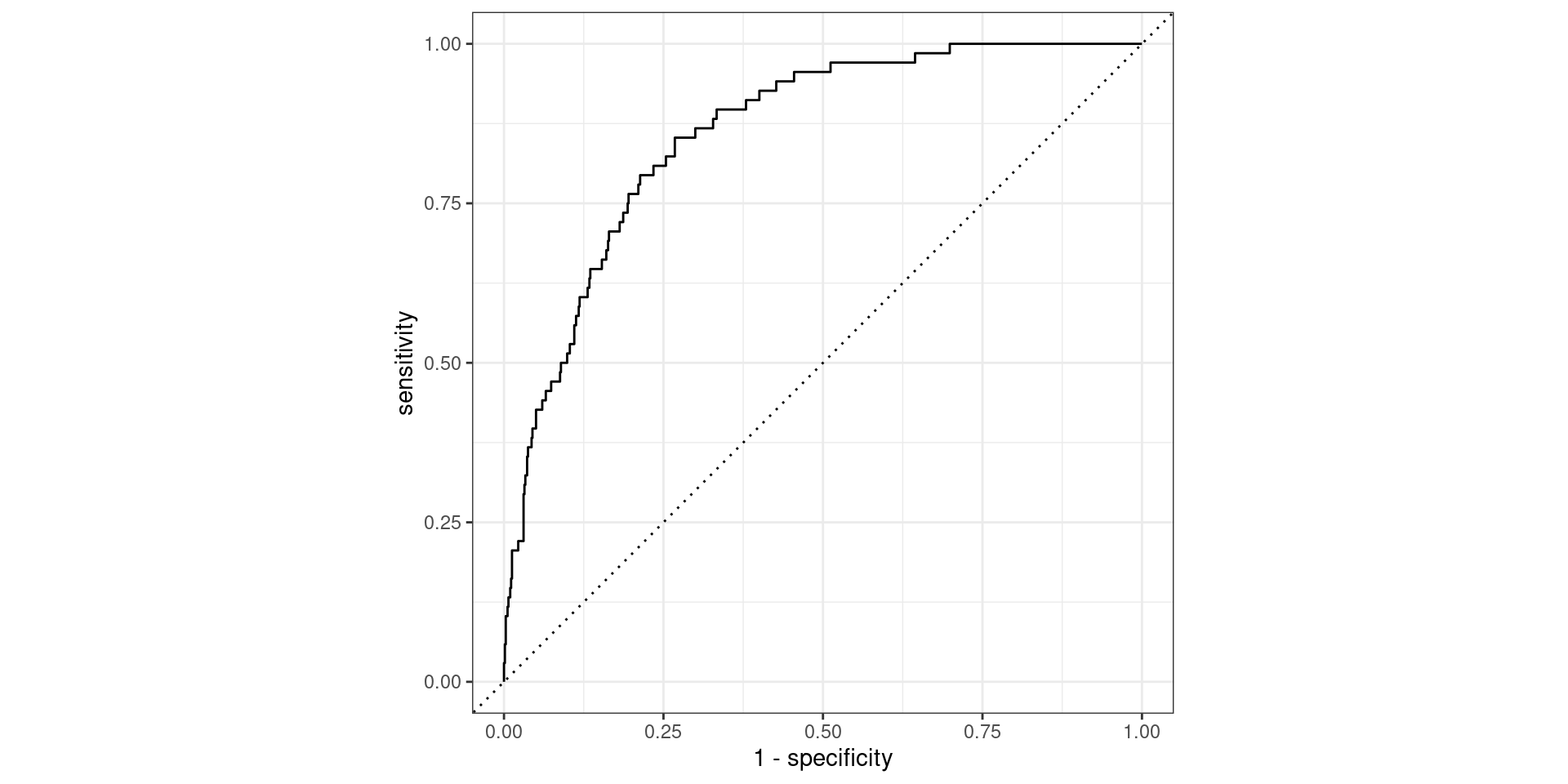
Evaluate the performance
# A tibble: 1 × 3
.metric .estimator .estimate
<chr> <chr> <dbl>
1 roc_auc binary 0.860This is better than our former model (without the feature engineering workflow), which had AUC = 0.857.
Making decisions
Cutoff probability: 0.5
Suppose we decide to label an email as spam if the model predicts the probability of spam to be more than 0.5. (That is the default.)
Confusion matrix:
cutoff_prob <- 0.5
email_pred |>
mutate(
spam = if_else(spam == 1, "Email is spam", "Email is not spam"),
spam_pred = if_else(.pred_1 > cutoff_prob, "Email labelled spam", "Email labelled not spam")
) |>
count(spam_pred, spam) |>
pivot_wider(names_from = spam_pred, values_from = n) |>
select(1,3,2) |> slice(c(2,1)) |> # reorder rows and cols to fit convention
knitr::kable()| spam | Email labelled spam | Email labelled not spam |
|---|---|---|
| Email is spam | 14 | 54 |
| Email is not spam | 10 | 707 |
Sensitivity: 14/(14+54) = 0.206
Specificity: 707/(707+10) = 0.986
Cutoff probability: 0.25
Suppose we decide to label an email as spam if the model predicts the probability of spam to be more than 0.25.
Confusion matrix:
| spam | Email labelled spam | Email labelled not spam |
|---|---|---|
| Email is spam | 32 | 36 |
| Email is not spam | 61 | 656 |
Sensitivity: 32/(32+36) = 0.471
Specificity: 656/(656 + 61) = 0.915
Cutoff probability: 0.75
Suppose we decide to label an email as spam if the model predicts the probability of spam to be more than 0.75.
Confusion matrix:
| spam | Email labelled spam | Email labelled not spam |
|---|---|---|
| Email is spam | 3 | 65 |
| Email is not spam | 1 | 716 |
Sensitivity: 3/(3+65) = 0.044
Specificity: 716/(716+1) = 0.999
Check our very first model
We make a new simple recipe and draw workflow and fitting re-using the same specified logisitc regression model email_mod.
simple_email_rec <- recipe(
spam ~ num_char, # formula
data = train_data # data to use for cataloguing names and types of variables
)
simple_email_pred <-
workflow() |>
add_model(email_mod) |>
add_recipe(simple_email_rec) |>
fit(data = train_data) |>
predict(test_data, type = "prob") |>
bind_cols(test_data |> select(spam,num_char,time))
simple_email_pred # A tibble: 785 × 5
.pred_0 .pred_1 spam num_char time
<dbl> <dbl> <fct> <dbl> <dttm>
1 0.889 0.111 0 4.84 2012-01-01 18:55:06
2 0.936 0.0644 0 15.1 2012-01-01 20:38:32
3 0.945 0.0547 0 18.0 2012-01-02 06:42:16
4 0.989 0.0113 0 45.8 2012-01-02 16:12:51
5 0.922 0.0784 0 11.4 2012-01-02 17:45:36
6 0.868 0.132 0 1.48 2012-01-02 22:55:03
7 0.933 0.0667 0 14.4 2012-01-03 02:07:17
8 0.864 0.136 0 0.978 2012-01-03 06:41:35
9 0.905 0.0953 0 7.79 2012-01-03 17:02:35
10 0.864 0.136 0 0.978 2012-01-03 12:14:51
# ℹ 775 more rowsEvaluate the performance
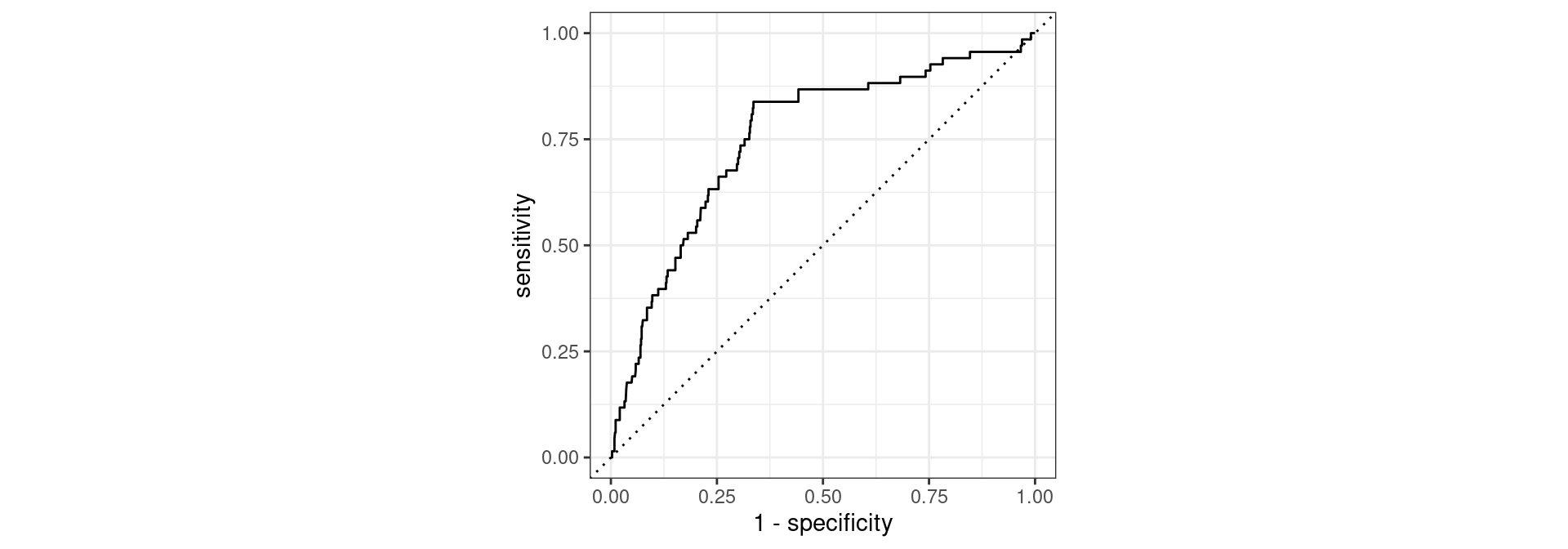
# A tibble: 1 × 3
.metric .estimator .estimate
<chr> <chr> <dbl>
1 roc_auc binary 0.753Conclusion: It is not as good compared to AUC 0.86
Predict Penguins’ Sex
Following all the steps introduced before:
set.seed(1)
# Put 80% of the data into the training set
peng_split <- initial_split(penguins, prop = 0.80)
# Create data frames for the two sets:
train_data_peng <- training(peng_split)
test_data_peng <- testing(peng_split)
# Create a recipe for the model
peng_rec <- recipe(
sex ~ .,
data = train_data_peng
) |>
step_naomit(all_predictors()) |>
step_dummy(all_nominal(), -all_outcomes()) |>
step_zv(all_predictors())
# Create a logistic regression model
peng_mod <- logistic_reg() |>
set_engine("glm")
# Create a workflow
peng_wflow <- workflow() |>
add_model(peng_mod) |>
add_recipe(peng_rec)
# Fitting model to training data
peng_fit <- peng_wflow |>
fit(data = train_data_peng)
# Make predictions for test data
peng_pred <- peng_fit |>
predict(test_data_peng, type = "prob") |>
bind_cols(test_data_peng)Penguins: Evaluate Performance
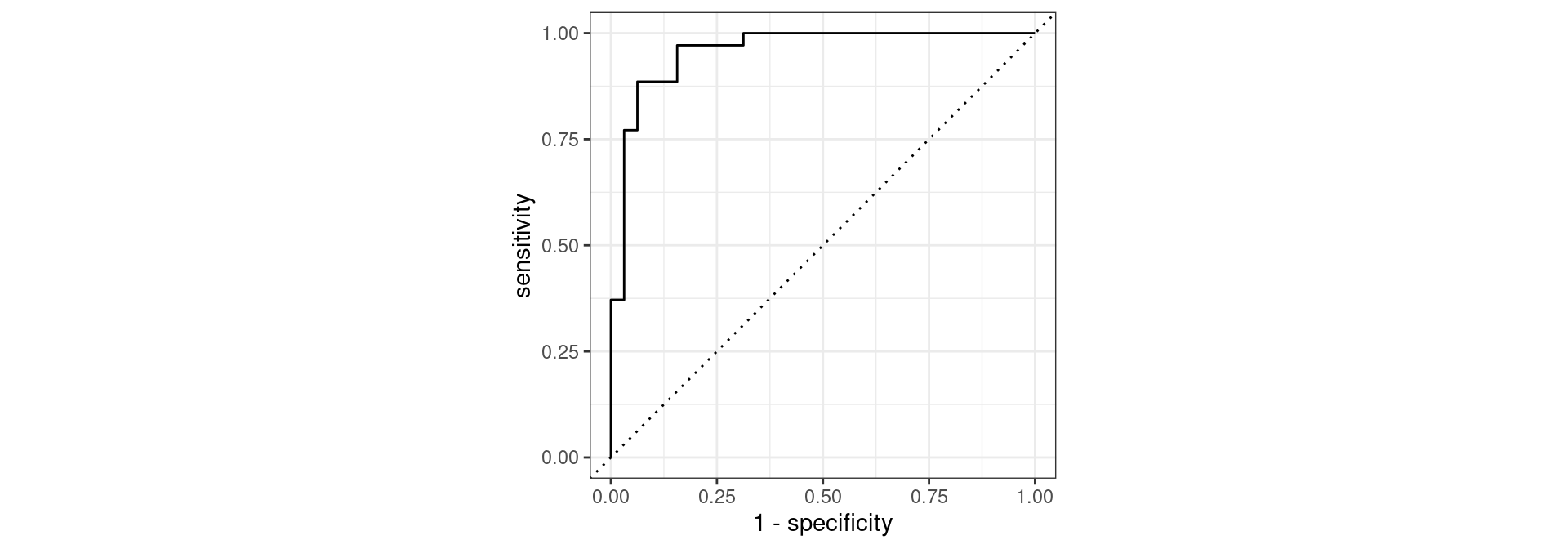
Penguins: Cutoff probability 0.5
Confusion matrix:
cutoff_prob <- 0.5
peng_pred |>
mutate(
sex = if_else(sex == "male", "Penguin is male", "Penguin is female"),
sex_pred = if_else(.pred_male > cutoff_prob, "Penguin labelled male", "Penguin labelled female")
) |>
count(sex_pred, sex) |>
pivot_wider(names_from = sex_pred, values_from = n) |>
select(1,3,2) |> slice(c(2,1)) |> # reorder rows and cols to fit convention
knitr::kable()| sex | Penguin labelled male | Penguin labelled female |
|---|---|---|
| Penguin is male | 31 | 4 |
| Penguin is female | 4 | 28 |
Sensitivity: 31/(31+4) = 0.886
Specificity: 28/(28+4) = 0.875
Penguins: Cutoff probability 0.25
Confusion matrix:
cutoff_prob <- 0.25
peng_pred |>
mutate(
sex = if_else(sex == "male", "Penguin is male", "Penguin is female"),
sex_pred = if_else(.pred_male > cutoff_prob, "Penguin labelled male", "Penguin labelled female")
) |>
count(sex_pred, sex) |>
pivot_wider(names_from = sex_pred, values_from = n) |>
select(1,3,2) |> slice(c(2,1)) |> # reorder rows and cols to fit convention
knitr::kable()| sex | Penguin labelled male | Penguin labelled female |
|---|---|---|
| Penguin is male | 34 | 1 |
| Penguin is female | 7 | 25 |
Sensitivity: 34/(34+1) = 0.971
Specificity: 25/(25+7) = 0.781
Penguins: Cutoff probability 0.75
Confusion matrix:
cutoff_prob <- 0.75
peng_pred |>
mutate(
sex = if_else(sex == "male", "Penguin is male", "Penguin is female"),
sex_pred = if_else(.pred_male > cutoff_prob, "Penguin labelled male", "Penguin labelled female")
) |>
count(sex_pred, sex) |>
pivot_wider(names_from = sex_pred, values_from = n) |>
select(1,3,2) |> slice(c(2,1)) |> # reorder rows and cols to fit convention
knitr::kable()| sex | Penguin labelled male | Penguin labelled female |
|---|---|---|
| Penguin is male | 29 | 6 |
| Penguin is female | 2 | 30 |
Sensitivity: 29/(29+6) = 0.829
Specificity: 30/(30+2) = 0.938